"afadin, adherens junction formation factor a"
ZFIN 
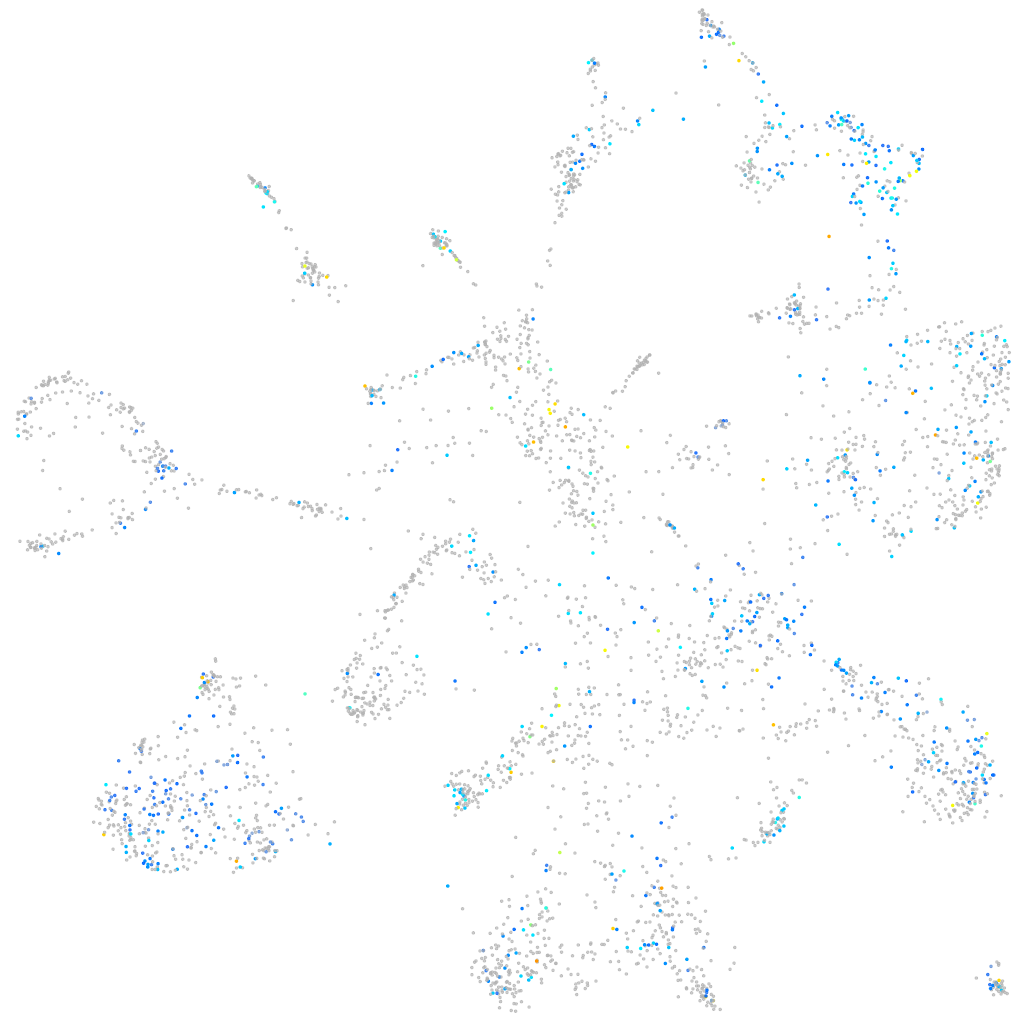
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sfrp5 | 0.188 | tpm1 | -0.112 |
| lmod2a | 0.183 | acta2 | -0.104 |
| CABZ01075068.1 | 0.176 | ak1 | -0.092 |
| mmel1 | 0.173 | gapdhs | -0.089 |
| XLOC-042222 | 0.164 | myl9a | -0.088 |
| tmem88b | 0.151 | ifitm1 | -0.087 |
| tbx5a | 0.147 | tagln | -0.086 |
| alpi.1 | 0.146 | apoa2 | -0.080 |
| tnni1b | 0.141 | csrp1b | -0.079 |
| alx4a | 0.139 | ptmaa | -0.076 |
| synpo2lb | 0.133 | pkma | -0.076 |
| gata5 | 0.132 | tmem119b | -0.076 |
| cdh2 | 0.132 | zgc:153704 | -0.072 |
| bnc2 | 0.128 | igfbp7 | -0.071 |
| nexn | 0.125 | ldha | -0.070 |
| bambia | 0.123 | mgll | -0.070 |
| podxl | 0.120 | eno1a | -0.070 |
| NC-002333.4 | 0.120 | ptx3a | -0.069 |
| jam2b | 0.118 | nme2b.2 | -0.068 |
| crb2a | 0.118 | bgnb | -0.067 |
| angpt1 | 0.118 | angptl5 | -0.065 |
| qkia | 0.118 | pgam1a | -0.065 |
| CABZ01072614.1 | 0.118 | aldocb | -0.065 |
| mdka | 0.114 | myl6 | -0.064 |
| nrip1b | 0.113 | zgc:158343 | -0.064 |
| NC-002333.17 | 0.112 | crip1 | -0.063 |
| zfpm1 | 0.112 | ckba | -0.063 |
| fabp11a | 0.112 | tpi1b | -0.062 |
| clec19a | 0.111 | cx43 | -0.062 |
| krt18a.1 | 0.111 | prss59.1 | -0.061 |
| jcada | 0.111 | prss1 | -0.061 |
| ftr82 | 0.110 | fbp1a | -0.060 |
| nr0b2a | 0.109 | fbp2 | -0.060 |
| cxadr | 0.109 | lmod1b | -0.060 |
| FITM1 (1 of many) | 0.109 | pgk1 | -0.060 |