amino-terminal enhancer of split [Source:ZFIN;Acc:ZDB-GENE-040426-1409]
ZFIN 
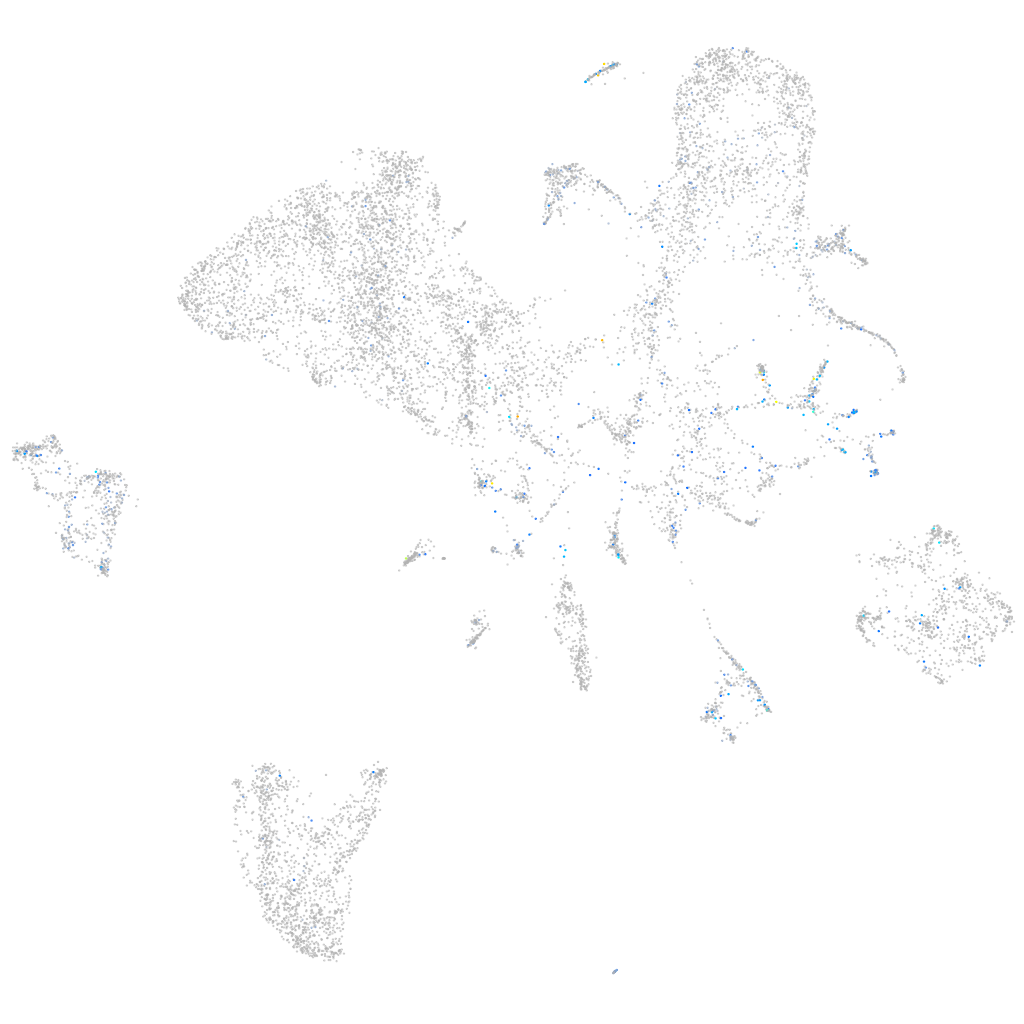
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.180 | gamt | -0.136 |
| neurod1 | 0.174 | gapdh | -0.131 |
| mir7a-1 | 0.170 | gatm | -0.129 |
| id4 | 0.166 | ahcy | -0.119 |
| cyth3b | 0.164 | bhmt | -0.117 |
| LOC110440045 | 0.164 | mat1a | -0.115 |
| pcsk1nl | 0.162 | fbp1b | -0.111 |
| LOC101885431 | 0.161 | apoa4b.1 | -0.107 |
| BX323801.1 | 0.161 | apoa1b | -0.106 |
| nkx2.2a | 0.157 | aqp12 | -0.104 |
| gng7 | 0.157 | agxtb | -0.103 |
| pax6b | 0.157 | apoc2 | -0.103 |
| vat1 | 0.156 | afp4 | -0.102 |
| ompa | 0.154 | apoc1 | -0.100 |
| gpc1a | 0.152 | aldob | -0.100 |
| gapdhs | 0.152 | glud1b | -0.096 |
| LOC110439817 | 0.150 | g6pca.2 | -0.096 |
| nrsn1 | 0.149 | pnp4b | -0.096 |
| jagn1a | 0.147 | apoa2 | -0.096 |
| spon1b | 0.147 | si:dkey-16p21.8 | -0.095 |
| scgn | 0.147 | agxta | -0.095 |
| tspan7b | 0.144 | grhprb | -0.094 |
| LOC110440108 | 0.144 | gpx4a | -0.093 |
| lysmd2 | 0.143 | serpina1 | -0.093 |
| tmsb | 0.141 | ces2 | -0.093 |
| kcnj19a | 0.140 | cx32.3 | -0.093 |
| LOC100537384 | 0.140 | serpina1l | -0.093 |
| rnasekb | 0.139 | LOC110437731 | -0.092 |
| tradv31.0 | 0.135 | dap | -0.092 |
| rims2a | 0.134 | apom | -0.092 |
| isl1 | 0.134 | hao1 | -0.092 |
| slc35g2a | 0.133 | abat | -0.092 |
| gpr22a | 0.133 | ftcd | -0.091 |
| capns1b | 0.133 | nupr1b | -0.091 |
| kcnj10a | 0.133 | scp2a | -0.091 |