AE binding protein 1
ZFIN 
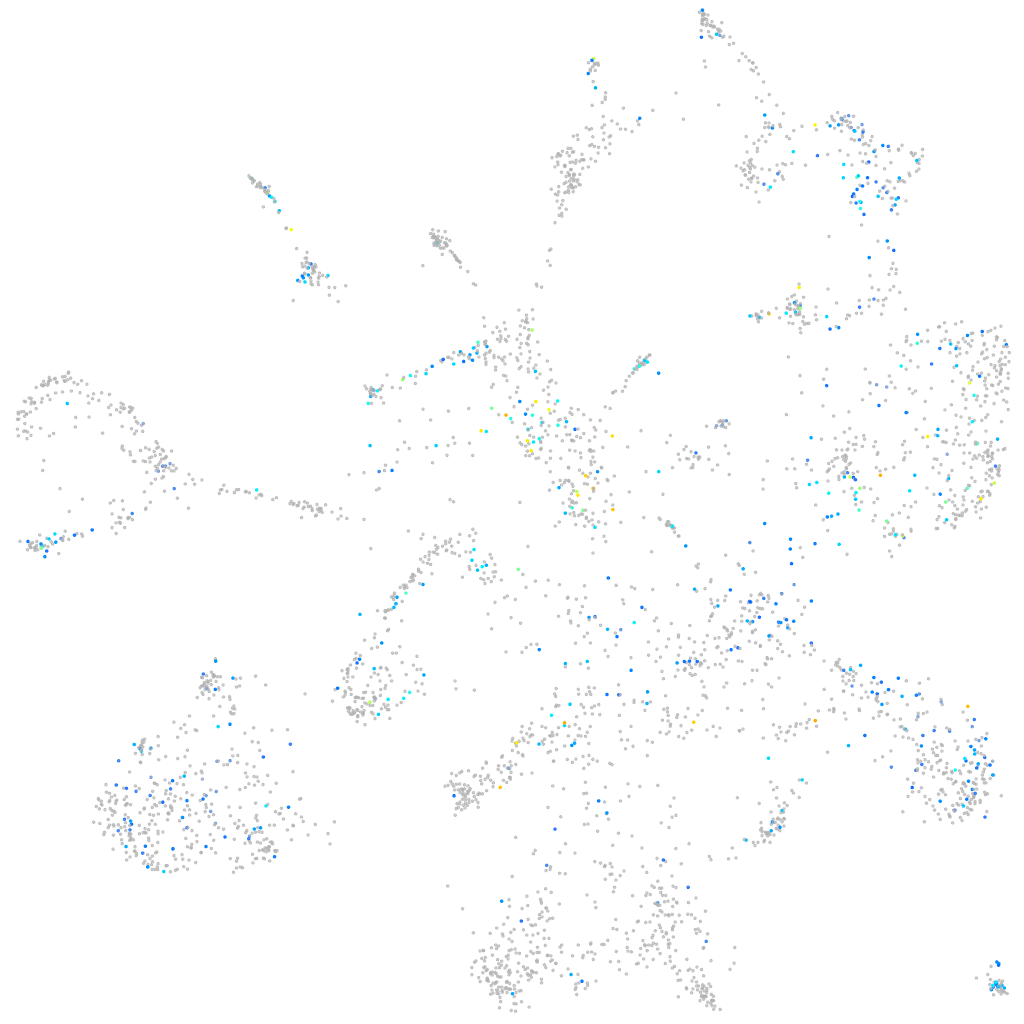
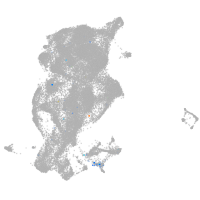
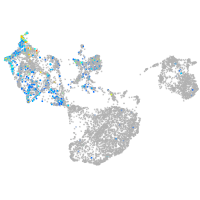
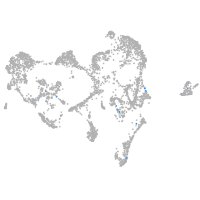
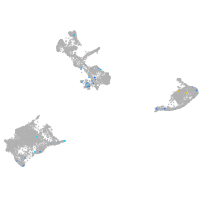
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksl1b | 0.144 | acta2 | -0.136 |
| si:dkey-238c7.16 | 0.127 | csrp1b | -0.122 |
| si:dkey-261h17.1 | 0.126 | myl9a | -0.120 |
| mdka | 0.121 | cald1b | -0.119 |
| nid1b | 0.119 | tagln | -0.118 |
| rbpms2b | 0.118 | desmb | -0.118 |
| si:ch211-129c21.1 | 0.117 | tpm2 | -0.113 |
| si:ch73-193i22.1 | 0.117 | mylkb | -0.112 |
| si:ch211-214p13.9 | 0.114 | cnn1b | -0.111 |
| sept2 | 0.114 | ckbb | -0.107 |
| si:ch211-39i22.1 | 0.112 | fhl1a | -0.101 |
| CR735102.1 | 0.111 | lmod1b | -0.099 |
| pgfb | 0.111 | myl6 | -0.094 |
| crabp2b | 0.111 | tpm1 | -0.093 |
| zeb2a | 0.111 | fbxl22 | -0.089 |
| si:ch211-286o17.1 | 0.108 | myo1ca | -0.089 |
| ednraa | 0.107 | rbm24a | -0.088 |
| tbx2b | 0.107 | si:ch211-137i24.10 | -0.086 |
| syn3 | 0.105 | tuba8l2 | -0.086 |
| thy1 | 0.105 | rgs2 | -0.085 |
| id1 | 0.103 | pgm5 | -0.083 |
| XLOC-043229 | 0.102 | vcla | -0.081 |
| zar1l | 0.100 | ckba | -0.081 |
| pon3.2 | 0.100 | myocd | -0.081 |
| gata2a | 0.099 | myh11a | -0.081 |
| cd248a | 0.099 | BX323087.1 | -0.079 |
| CR407547.1 | 0.097 | itpka | -0.079 |
| XLOC-024493 | 0.097 | akap7 | -0.078 |
| mfap2 | 0.097 | smtna | -0.076 |
| entpd1 | 0.097 | acta1a | -0.076 |
| phactr4b | 0.096 | acta1b | -0.076 |
| mmp2 | 0.096 | pdlim3a | -0.075 |
| CR392341.3 | 0.096 | il13ra2 | -0.075 |
| chadlb | 0.095 | gucy1a1 | -0.074 |
| glt8d2 | 0.095 | pnp4a | -0.074 |