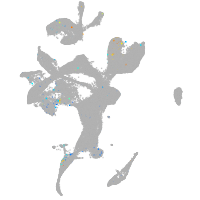
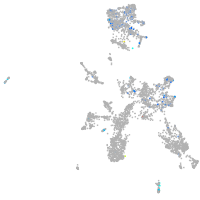
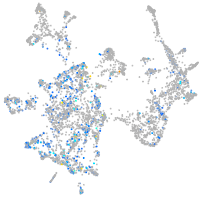
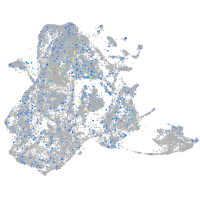
adenylate cyclase 7
ZFIN 
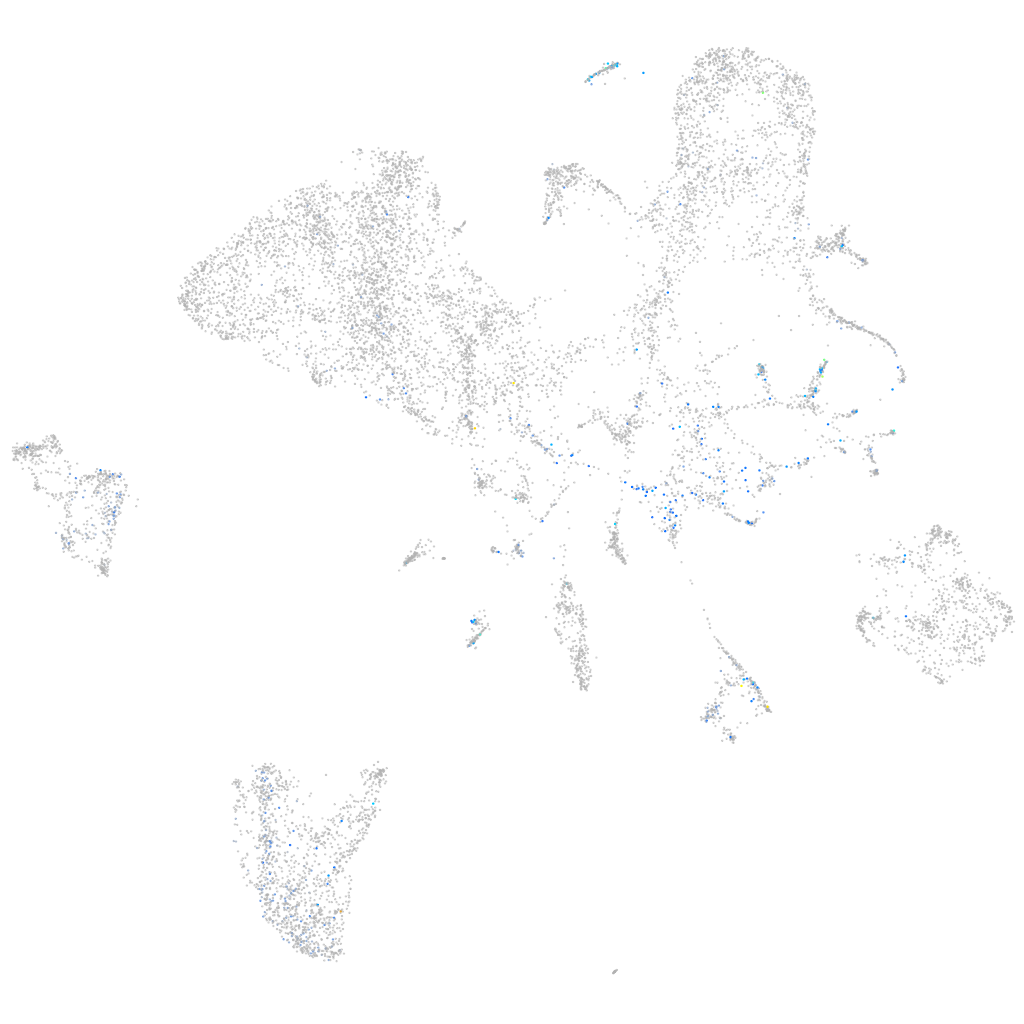
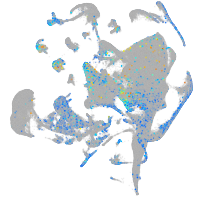
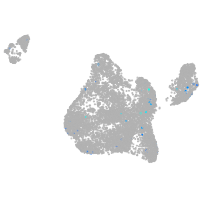
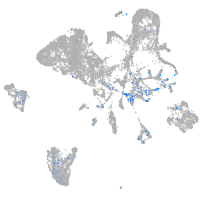
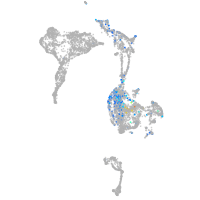
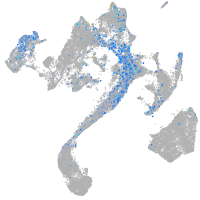
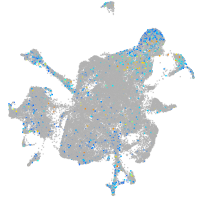
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.169 | gapdh | -0.120 |
| LOC101883054 | 0.167 | apoa4b.1 | -0.114 |
| XLOC-024390 | 0.163 | apoc2 | -0.112 |
| adcyap1a | 0.147 | afp4 | -0.110 |
| insm1b | 0.146 | fbp1b | -0.110 |
| fev | 0.146 | gstt1a | -0.107 |
| efcab2 | 0.143 | scp2a | -0.105 |
| CABZ01025162.1 | 0.141 | aldob | -0.104 |
| trpa1b | 0.141 | suclg1 | -0.104 |
| LO018188.1 | 0.139 | si:dkey-16p21.8 | -0.104 |
| zgc:101731 | 0.138 | gamt | -0.101 |
| CU914622.1 | 0.136 | apoc1 | -0.101 |
| HTRA2 (1 of many).6 | 0.134 | apoa1b | -0.100 |
| vat1 | 0.134 | ahcy | -0.100 |
| egr4 | 0.131 | eno3 | -0.098 |
| znf1085 | 0.131 | apoa2 | -0.095 |
| dnah7 | 0.131 | apobb.1 | -0.095 |
| c2cd4a | 0.131 | gpx4a | -0.094 |
| LOC557811 | 0.129 | mgst1.2 | -0.093 |
| tmtops3b | 0.129 | suclg2 | -0.093 |
| slc7a14a | 0.128 | nipsnap3a | -0.092 |
| adgrg4b | 0.126 | apom | -0.092 |
| CR383676.1 | 0.125 | sult2st2 | -0.092 |
| si:ch211-238e22.5 | 0.124 | tdo2a | -0.091 |
| hpcal1 | 0.124 | hao1 | -0.091 |
| id4 | 0.124 | ftcd | -0.091 |
| LOC103909163 | 0.123 | slc27a2a | -0.091 |
| syt1a | 0.122 | g6pca.2 | -0.090 |
| gabra2a | 0.122 | rbp2b | -0.090 |
| XLOC-030409 | 0.122 | rbp4 | -0.090 |
| pcsk1nl | 0.121 | ces2 | -0.090 |
| dll4 | 0.121 | ugt1a7 | -0.090 |
| nkx2.2a | 0.120 | serpina1l | -0.090 |
| slc35g2a | 0.119 | zgc:123103 | -0.089 |
| neurod1 | 0.118 | serpina1 | -0.089 |