adenosine deaminase RNA specific
ZFIN 
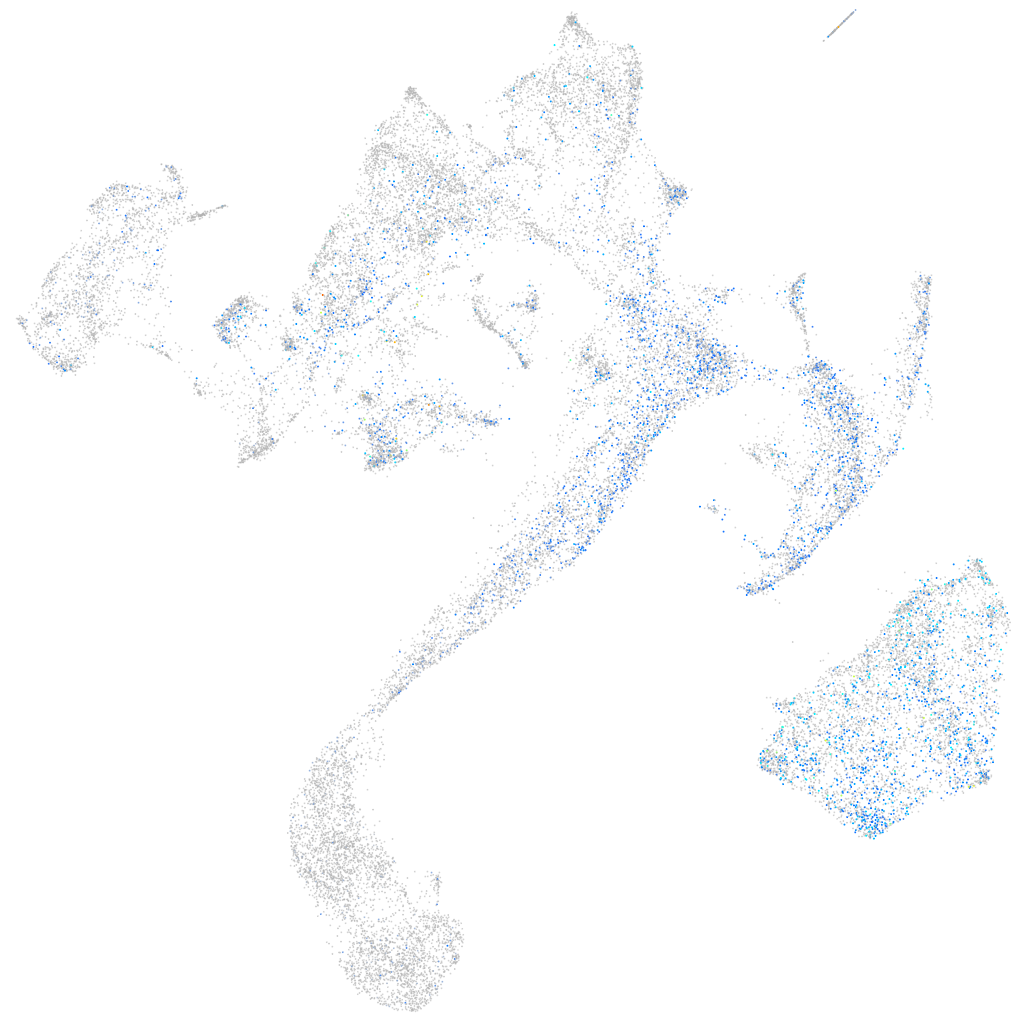
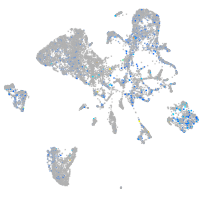
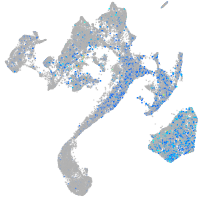
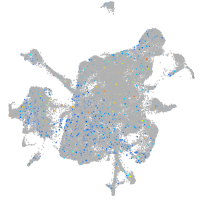
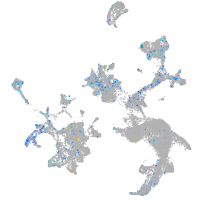
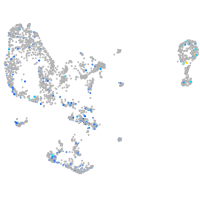
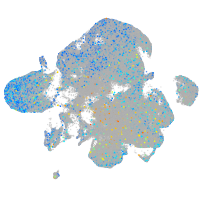
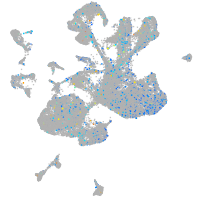
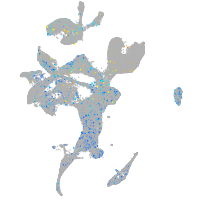
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ncl | 0.212 | rpl37 | -0.156 |
| nop58 | 0.206 | actc1b | -0.153 |
| fbl | 0.204 | ckma | -0.143 |
| nop56 | 0.203 | zgc:114188 | -0.143 |
| dkc1 | 0.202 | ckmb | -0.142 |
| npm1a | 0.198 | rps10 | -0.139 |
| syncrip | 0.198 | ak1 | -0.138 |
| hnrnpa1b | 0.196 | bhmt | -0.137 |
| ppig | 0.196 | mylpfa | -0.137 |
| hnrnpabb | 0.196 | tnnc2 | -0.136 |
| anp32e | 0.195 | atp2a1 | -0.135 |
| hnrnpub | 0.193 | rps17 | -0.135 |
| hmga1a | 0.192 | fabp3 | -0.134 |
| nucks1a | 0.191 | pvalb1 | -0.133 |
| hnrnpa0b | 0.190 | vdac3 | -0.131 |
| bms1 | 0.190 | acta1b | -0.131 |
| khdrbs1a | 0.190 | pvalb2 | -0.130 |
| nop2 | 0.189 | neb | -0.129 |
| seta | 0.189 | mylz3 | -0.129 |
| rbm4.3 | 0.188 | aldoab | -0.129 |
| cirbpa | 0.188 | tpi1b | -0.129 |
| setb | 0.188 | tpma | -0.129 |
| top1l | 0.188 | si:ch73-367p23.2 | -0.129 |
| marcksb | 0.188 | ldb3b | -0.128 |
| cx43.4 | 0.187 | eef1da | -0.127 |
| hnrnpa1a | 0.184 | eno3 | -0.127 |
| snrnp70 | 0.184 | tnnt3a | -0.126 |
| hnrnpaba | 0.183 | actn3a | -0.125 |
| hnrnpm | 0.183 | smyd1a | -0.125 |
| snrpb | 0.182 | nme2b.2 | -0.123 |
| gar1 | 0.182 | CABZ01078594.1 | -0.123 |
| brd3a | 0.182 | actn3b | -0.122 |
| cbx3a | 0.181 | myom1a | -0.122 |
| srsf1a | 0.181 | tnnt3b | -0.121 |
| srrm2 | 0.181 | hhatla | -0.121 |