ADAM metallopeptidase domain 19b
ZFIN 
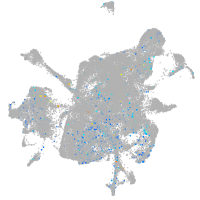
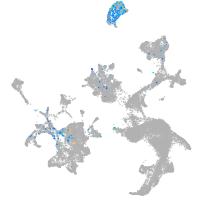
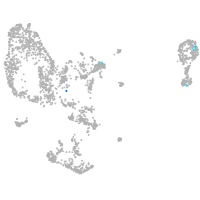
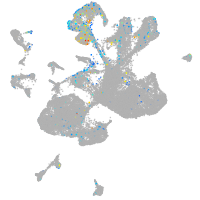
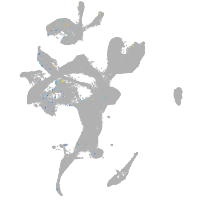
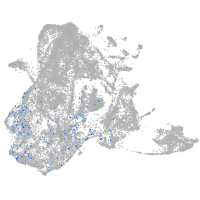
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npvf | 0.396 | eno3 | -0.033 |
| tmem200a | 0.356 | aldh6a1 | -0.032 |
| LOC103908925 | 0.340 | si:dkey-16p21.8 | -0.032 |
| LOC103909512 | 0.337 | cebpd | -0.032 |
| LOC101885341 | 0.329 | eef1da | -0.031 |
| pappab | 0.328 | aldob | -0.029 |
| cacna2d2b | 0.318 | abat | -0.029 |
| si:ch211-241b2.4 | 0.314 | rps28 | -0.029 |
| CABZ01088229.1 | 0.272 | fbp1b | -0.028 |
| brinp3a.2 | 0.268 | acaa2 | -0.028 |
| sgk494a | 0.266 | COX7A2 (1 of many) | -0.028 |
| mhc1zda | 0.262 | ndufa4l | -0.028 |
| ebf1a | 0.260 | rpl39 | -0.028 |
| sybu | 0.260 | pgam1a | -0.028 |
| mtus2a | 0.256 | chchd10 | -0.027 |
| barhl1a | 0.250 | gamt | -0.027 |
| pcdh1g29 | 0.246 | nupr1b | -0.027 |
| LOC100537342 | 0.242 | gstm.1 | -0.027 |
| cacng8b | 0.239 | mat1a | -0.027 |
| si:ch211-119e14.1 | 0.233 | nipsnap3a | -0.027 |
| trarg1a | 0.232 | gatm | -0.027 |
| lrch2 | 0.231 | echs1 | -0.027 |
| ksr2 | 0.229 | cyb5a | -0.027 |
| barhl1b | 0.225 | etfb | -0.027 |
| LOC110438534 | 0.221 | COX3 | -0.027 |
| st8sia5 | 0.218 | cx32.3 | -0.027 |
| TMEM164 | 0.216 | mdh1aa | -0.027 |
| st6galnac5a | 0.214 | glud1b | -0.027 |
| si:dkey-240n22.6 | 0.214 | gapdh | -0.026 |
| si:dkey-79d12.6 | 0.213 | agxta | -0.026 |
| add2 | 0.212 | cat | -0.026 |
| thsd7aa | 0.209 | agxtb | -0.026 |
| ttbk1a | 0.209 | aldh7a1 | -0.026 |
| rps6kl1 | 0.207 | grhprb | -0.026 |
| gabrp | 0.202 | uqcrq | -0.026 |