"acid phosphatase 6, lysophosphatidic"
ZFIN 
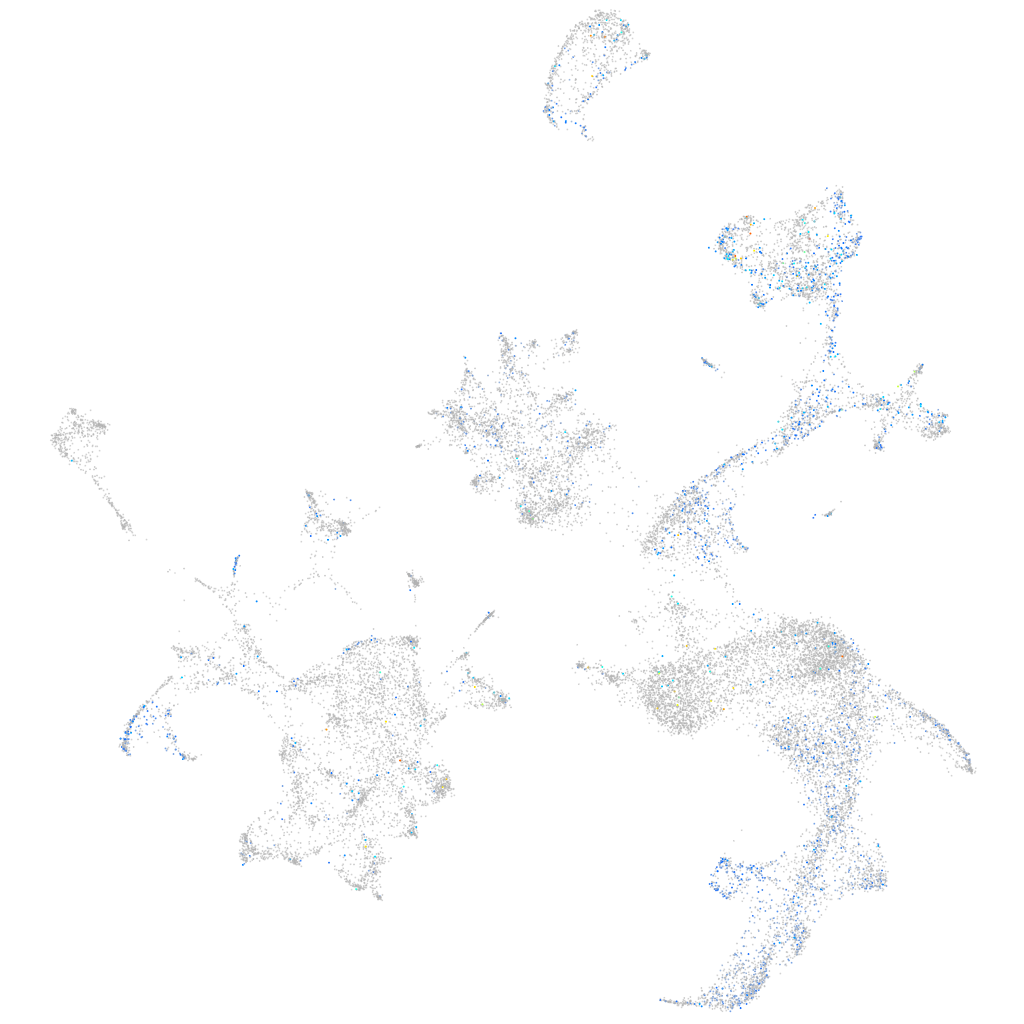
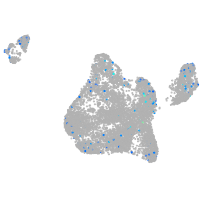
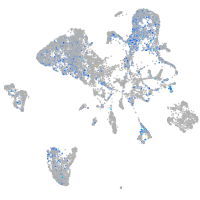
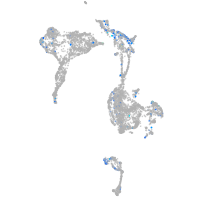
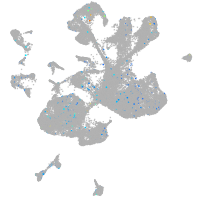
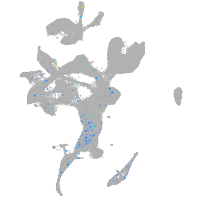
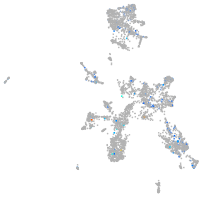
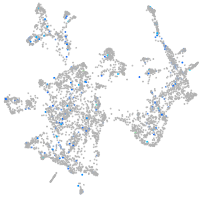
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cd83 | 0.157 | si:ch211-250g4.3 | -0.083 |
| cycsb | 0.148 | krt18a.1 | -0.075 |
| atp5mc1 | 0.143 | cdh5 | -0.072 |
| suclg1 | 0.143 | creg1 | -0.072 |
| cox17 | 0.138 | dap1b | -0.070 |
| shmt1 | 0.137 | akap12b | -0.070 |
| ppifb | 0.132 | clec14a | -0.065 |
| rac2 | 0.132 | jpt1b | -0.064 |
| coro1a | 0.131 | id1 | -0.063 |
| ptges3b | 0.127 | nova2 | -0.063 |
| cxcr3.2 | 0.127 | tmem88a | -0.062 |
| prelid3b | 0.127 | she | -0.062 |
| zgc:56493 | 0.126 | pecam1 | -0.061 |
| laptm5 | 0.126 | igfbp7 | -0.060 |
| tcp1 | 0.125 | krt8 | -0.060 |
| atp5f1b | 0.125 | etv2 | -0.059 |
| cct7 | 0.125 | si:ch211-156j16.1 | -0.058 |
| abce1 | 0.124 | plvapb | -0.058 |
| phb2b | 0.124 | tpm4a | -0.057 |
| kars | 0.123 | si:ch211-248e11.2 | -0.057 |
| ptpn6 | 0.123 | cpn1 | -0.056 |
| nars | 0.123 | tie1 | -0.055 |
| lgals2a | 0.122 | sox7 | -0.055 |
| c1qbp | 0.122 | tspan4b | -0.055 |
| atp5f1c | 0.122 | zgc:92606 | -0.055 |
| hspd1 | 0.121 | pmp22a | -0.055 |
| CU499330.1 | 0.121 | efnb2b | -0.054 |
| tnfb | 0.120 | egfl7 | -0.054 |
| higd1a | 0.120 | akt1s1 | -0.053 |
| eif4a1a | 0.120 | ldb2a | -0.053 |
| arpc1b | 0.119 | LOC108179634 | -0.053 |
| stoml3b | 0.119 | hmgb3a | -0.052 |
| pitpnaa | 0.119 | col4a1 | -0.052 |
| spi1a | 0.118 | cldn5b | -0.052 |
| ndufa10 | 0.118 | sox18 | -0.052 |