"acid phosphatase 5b, tartrate resistant"
ZFIN 
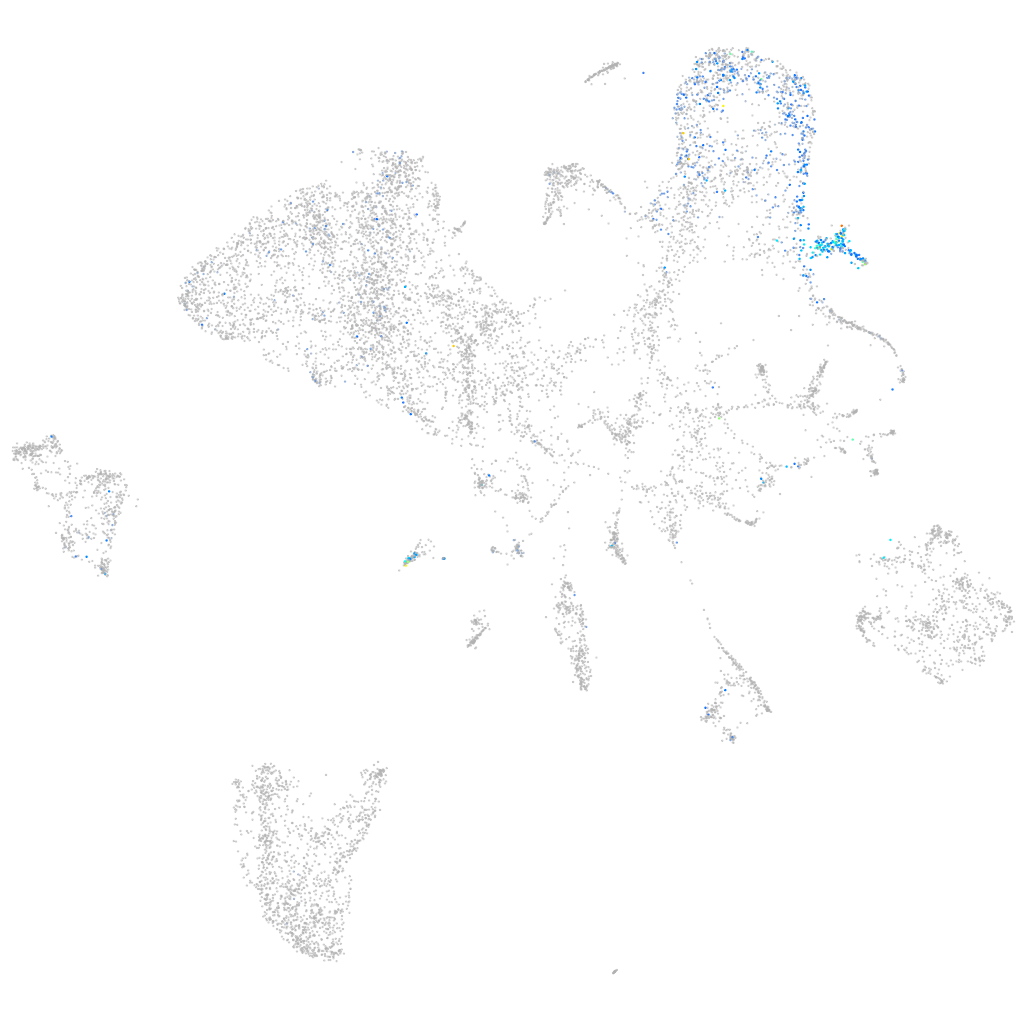
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctsl.1 | 0.520 | fabp3 | -0.157 |
| epdl2 | 0.503 | aqp12 | -0.153 |
| si:dkey-28n18.9 | 0.466 | bhmt | -0.151 |
| ctsbb | 0.439 | agxtb | -0.122 |
| IZUMO1R | 0.438 | apoc1 | -0.118 |
| LOC100537272 | 0.436 | kng1 | -0.117 |
| lrp2b | 0.436 | wu:fj16a03 | -0.115 |
| cubn | 0.417 | ttc36 | -0.112 |
| amn | 0.417 | tfa | -0.112 |
| grn1 | 0.410 | serpinf2b | -0.108 |
| dab2 | 0.407 | vtnb | -0.108 |
| slc15a2 | 0.405 | grhprb | -0.108 |
| tfeb | 0.405 | apoa2 | -0.107 |
| CR381686.5 | 0.405 | serpina1 | -0.107 |
| sptbn5 | 0.402 | serpina1l | -0.107 |
| fabp6 | 0.402 | cfhl4 | -0.106 |
| si:ch211-196f2.3 | 0.399 | ppdpfa | -0.106 |
| xirp1 | 0.390 | rbp2b | -0.106 |
| rab32b | 0.383 | hpda | -0.104 |
| naga | 0.383 | hao1 | -0.104 |
| tspan34 | 0.380 | fgg | -0.104 |
| cdaa | 0.372 | uox | -0.104 |
| dpep1 | 0.370 | cpn1 | -0.104 |
| NAT16 (1 of many) | 0.368 | zgc:123103 | -0.103 |
| si:ch211-139a5.9 | 0.367 | apom | -0.103 |
| zanl | 0.367 | ambp | -0.101 |
| si:dkey-283b1.6 | 0.364 | fgb | -0.100 |
| tmigd1 | 0.361 | fetub | -0.100 |
| enpep | 0.361 | ces2 | -0.099 |
| AL831745.1 | 0.358 | rbp4 | -0.099 |
| scpep1 | 0.355 | pygl | -0.099 |
| p2rx4a | 0.351 | lcn15 | -0.099 |
| si:dkey-194e6.1 | 0.351 | f2 | -0.098 |
| slc6a18 | 0.350 | fabp10a | -0.098 |
| slc10a2 | 0.350 | fga | -0.098 |