acyl-CoA dehydrogenase very long chain
ZFIN 
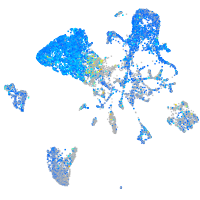
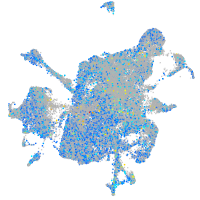
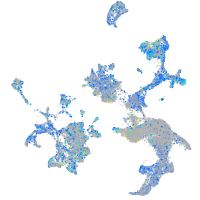
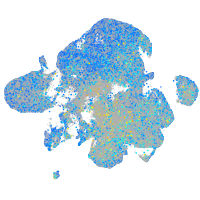
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX000438.2 | 0.247 | si:ch73-23l24.1 | -0.151 |
| bik | 0.246 | cnmd | -0.150 |
| ponzr5 | 0.232 | col2a1a | -0.149 |
| si:dkey-72l14.3 | 0.231 | col9a1b | -0.146 |
| epdl1 | 0.229 | zgc:113232 | -0.145 |
| gpa33a | 0.221 | col9a2 | -0.145 |
| spint2 | 0.216 | col11a1a | -0.143 |
| cd9b | 0.216 | col9a3 | -0.142 |
| CU855878.1 | 0.214 | zmp:0000000760 | -0.142 |
| zgc:85932 | 0.207 | cmn | -0.139 |
| cracr2b | 0.206 | col11a2 | -0.134 |
| mctp2b | 0.205 | ucmab | -0.132 |
| fuca1.1 | 0.202 | epyc | -0.127 |
| zgc:101744 | 0.201 | ppib | -0.127 |
| epcam | 0.201 | matn3a | -0.116 |
| si:ch211-79k12.1 | 0.200 | col8a1a | -0.115 |
| cldn7b | 0.198 | krt5 | -0.110 |
| LOC110439414 | 0.198 | sparc | -0.109 |
| prg4b | 0.197 | si:dkey-31g6.6 | -0.108 |
| tmprss9 | 0.197 | col2a1b | -0.106 |
| si:ch211-105j21.9 | 0.195 | ctgfa | -0.106 |
| nipal4 | 0.195 | krt18a.1 | -0.105 |
| sqstm1 | 0.193 | loxl1 | -0.100 |
| si:ch211-264f5.6 | 0.193 | thbs1b | -0.100 |
| hpgd | 0.193 | slc7a8a | -0.096 |
| cyp17a1 | 0.192 | snorc | -0.095 |
| tmem176l.4 | 0.192 | susd5 | -0.095 |
| il34 | 0.191 | plod1a | -0.095 |
| ngfrb | 0.190 | matn4 | -0.095 |
| cox4i2 | 0.190 | si:ch211-13f8.2 | -0.092 |
| glrx | 0.189 | selenom | -0.091 |
| nnt | 0.188 | si:dkey-6n6.1 | -0.091 |
| ildr1a | 0.188 | chad | -0.091 |
| ak1 | 0.187 | tgfbi | -0.091 |
| mt2 | 0.187 | zgc:158423 | -0.091 |