acyl-CoA dehydrogenase long chain
ZFIN 
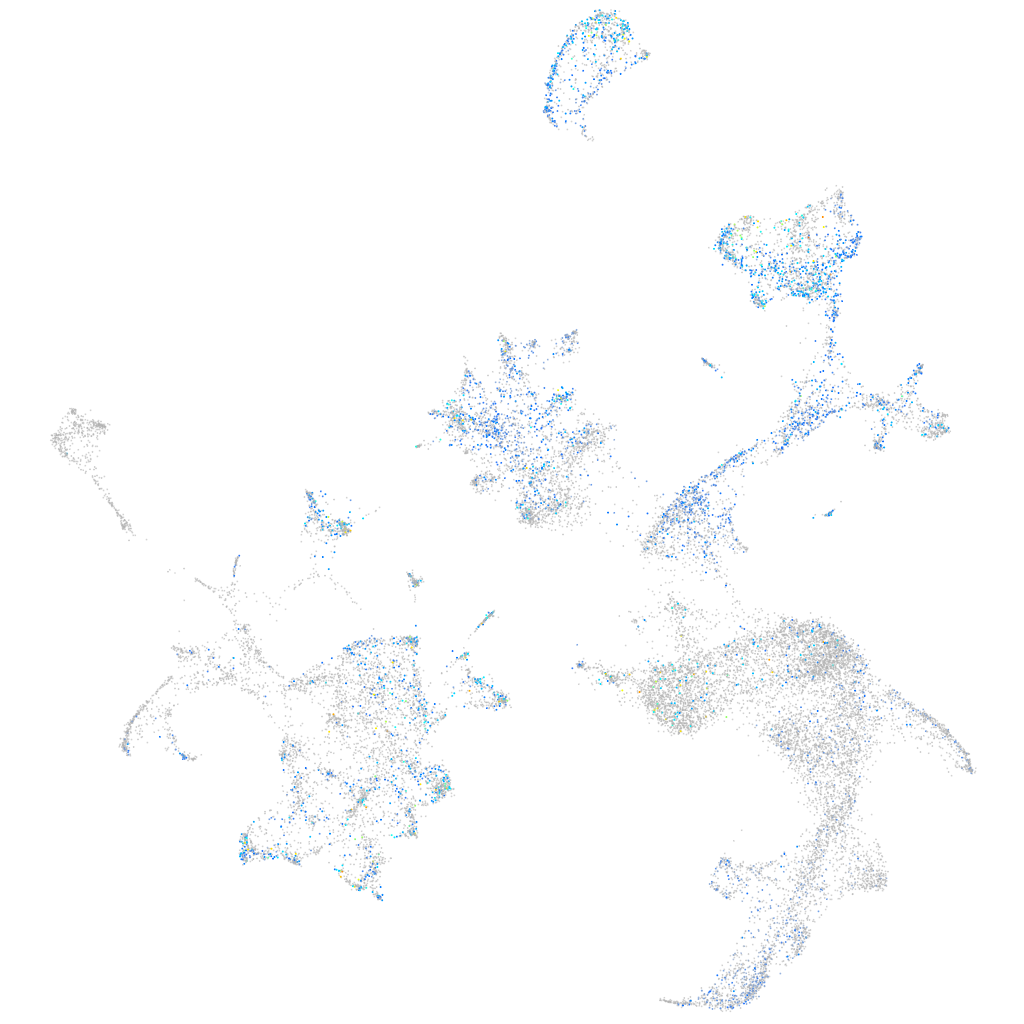
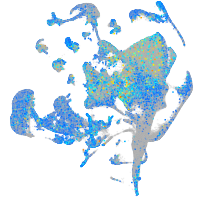
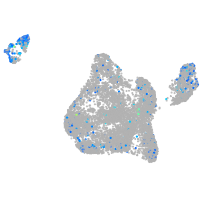
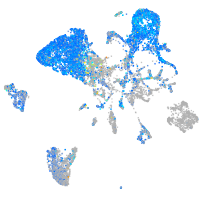
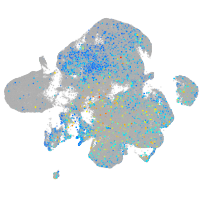
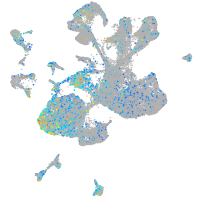
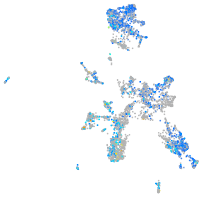
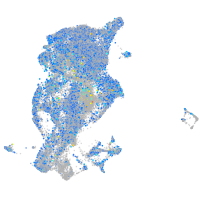
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.221 | hbae1.3 | -0.171 |
| nme2b.1 | 0.219 | hbae3 | -0.167 |
| eno3 | 0.219 | hbbe1.3 | -0.167 |
| arpc1b | 0.218 | hbae1.1 | -0.166 |
| cotl1 | 0.217 | cahz | -0.155 |
| cd63 | 0.214 | hbbe1.1 | -0.155 |
| cebpb | 0.213 | hbbe2 | -0.153 |
| laptm5 | 0.212 | hbbe1.2 | -0.149 |
| actb2 | 0.209 | alas2 | -0.147 |
| xbp1 | 0.204 | hemgn | -0.134 |
| arpc2 | 0.203 | fth1a | -0.131 |
| coro1a | 0.203 | si:ch211-250g4.3 | -0.130 |
| ctss2.1 | 0.202 | nt5c2l1 | -0.130 |
| pfn1 | 0.201 | slc4a1a | -0.129 |
| arpc3 | 0.201 | zgc:163057 | -0.128 |
| zgc:162730 | 0.200 | creg1 | -0.120 |
| ppiab | 0.200 | hbbe3 | -0.117 |
| nfkbiab | 0.199 | epb41b | -0.114 |
| pkma | 0.199 | si:ch211-207c6.2 | -0.112 |
| rac2 | 0.199 | blvrb | -0.112 |
| eef1da | 0.199 | tmod4 | -0.110 |
| crip1 | 0.199 | nmt1b | -0.100 |
| gsto2 | 0.198 | zgc:56095 | -0.100 |
| pgd | 0.196 | tspo | -0.099 |
| cap1 | 0.195 | plac8l1 | -0.096 |
| arhgdig | 0.195 | si:ch211-227m13.1 | -0.095 |
| actb1 | 0.195 | hbae5 | -0.092 |
| cyba | 0.195 | sptb | -0.088 |
| tmsb4x | 0.194 | si:dkey-240n22.3 | -0.084 |
| arpc5b | 0.193 | CR293511.1 | -0.081 |
| arpc4 | 0.192 | tfr1a | -0.079 |
| zgc:64051 | 0.191 | add2 | -0.079 |
| GCA | 0.191 | rfesd | -0.077 |
| capgb | 0.190 | uros | -0.077 |
| lasp1 | 0.189 | ucp3 | -0.075 |