"acyl-CoA dehydrogenase family, member 11"
ZFIN 
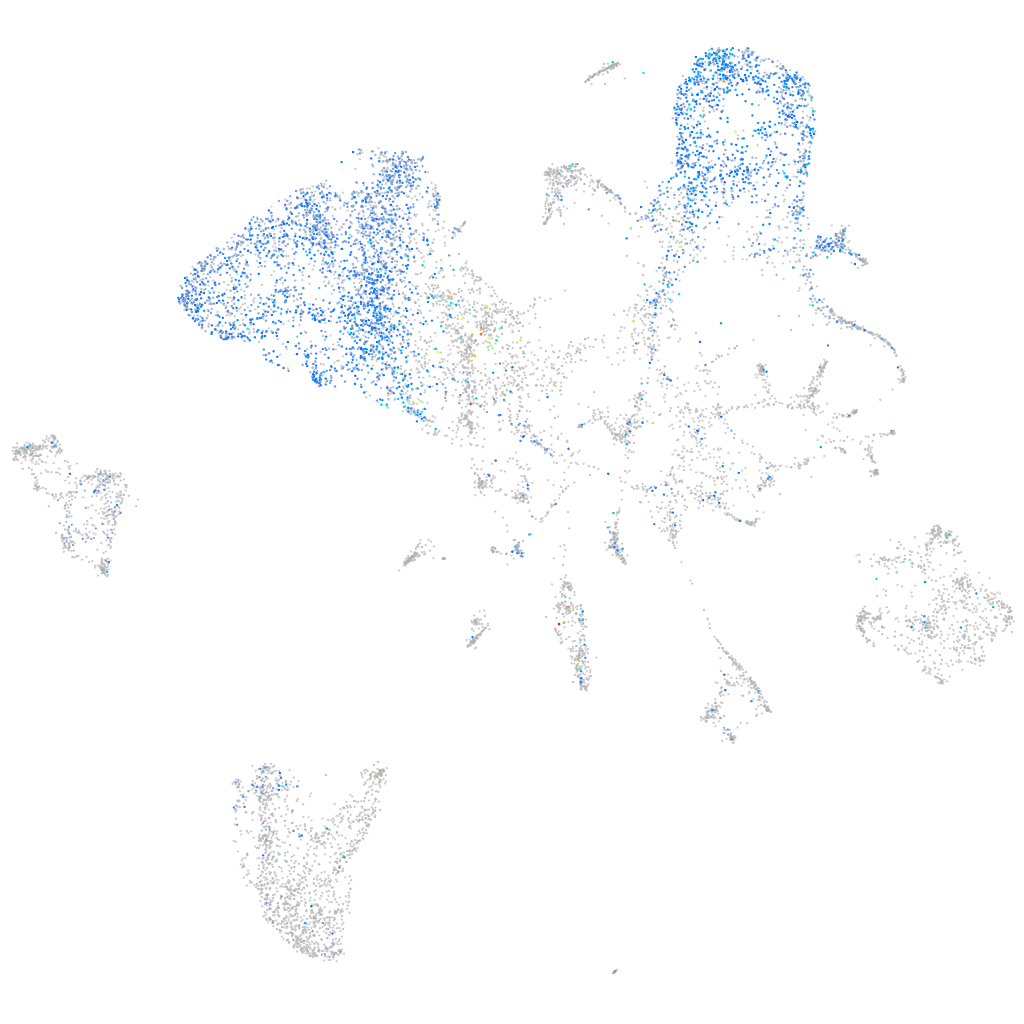
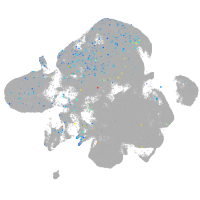
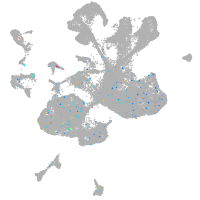
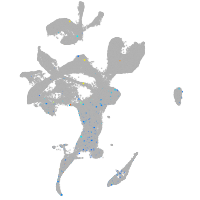
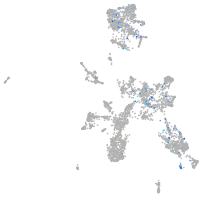
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ugt1a7 | 0.464 | slc38a5b | -0.223 |
| glud1b | 0.460 | hmgb3a | -0.215 |
| scp2a | 0.450 | rtn1a | -0.213 |
| mttp | 0.447 | epcam | -0.205 |
| sult2st2 | 0.445 | marcksl1b | -0.193 |
| srd5a2a | 0.442 | hmgn6 | -0.190 |
| rdh1 | 0.436 | jpt1b | -0.190 |
| dhrs9 | 0.429 | marcksb | -0.189 |
| apoa4b.1 | 0.427 | glulb | -0.189 |
| cyp8b1 | 0.426 | si:ch211-195b11.3 | -0.185 |
| gcshb | 0.425 | si:ch211-222l21.1 | -0.182 |
| apoc2 | 0.424 | ctrl | -0.182 |
| gpx4a | 0.423 | cldnb | -0.179 |
| gstr | 0.421 | hmgb1b | -0.178 |
| gstt1a | 0.419 | ywhaz | -0.176 |
| fdx1 | 0.417 | zgc:112160 | -0.173 |
| mdh1aa | 0.417 | prss59.1 | -0.172 |
| fbp1b | 0.412 | cldn7b | -0.171 |
| abcc2 | 0.411 | ctrb1 | -0.171 |
| sod1 | 0.410 | cpa5 | -0.171 |
| suclg1 | 0.408 | ela2 | -0.170 |
| apobb.1 | 0.408 | prss59.2 | -0.170 |
| sdr16c5b | 0.406 | cpb1 | -0.170 |
| eno3 | 0.405 | ela2l | -0.170 |
| slc37a4a | 0.405 | CELA1 (1 of many) | -0.170 |
| gapdh | 0.402 | sycn.2 | -0.169 |
| ugt5b4 | 0.400 | fep15 | -0.169 |
| cyp2y3 | 0.399 | ela3l | -0.169 |
| dgat2 | 0.398 | c6ast4 | -0.169 |
| slco1d1 | 0.395 | apoda.2 | -0.168 |
| prdx2 | 0.394 | erp27 | -0.168 |
| clic5b | 0.394 | id1 | -0.167 |
| haao | 0.393 | cel.1 | -0.167 |
| creb3l3a | 0.392 | si:ch211-240l19.5 | -0.167 |
| acaa1 | 0.389 | cx43.4 | -0.166 |