abhydrolase domain containing 15a
ZFIN 
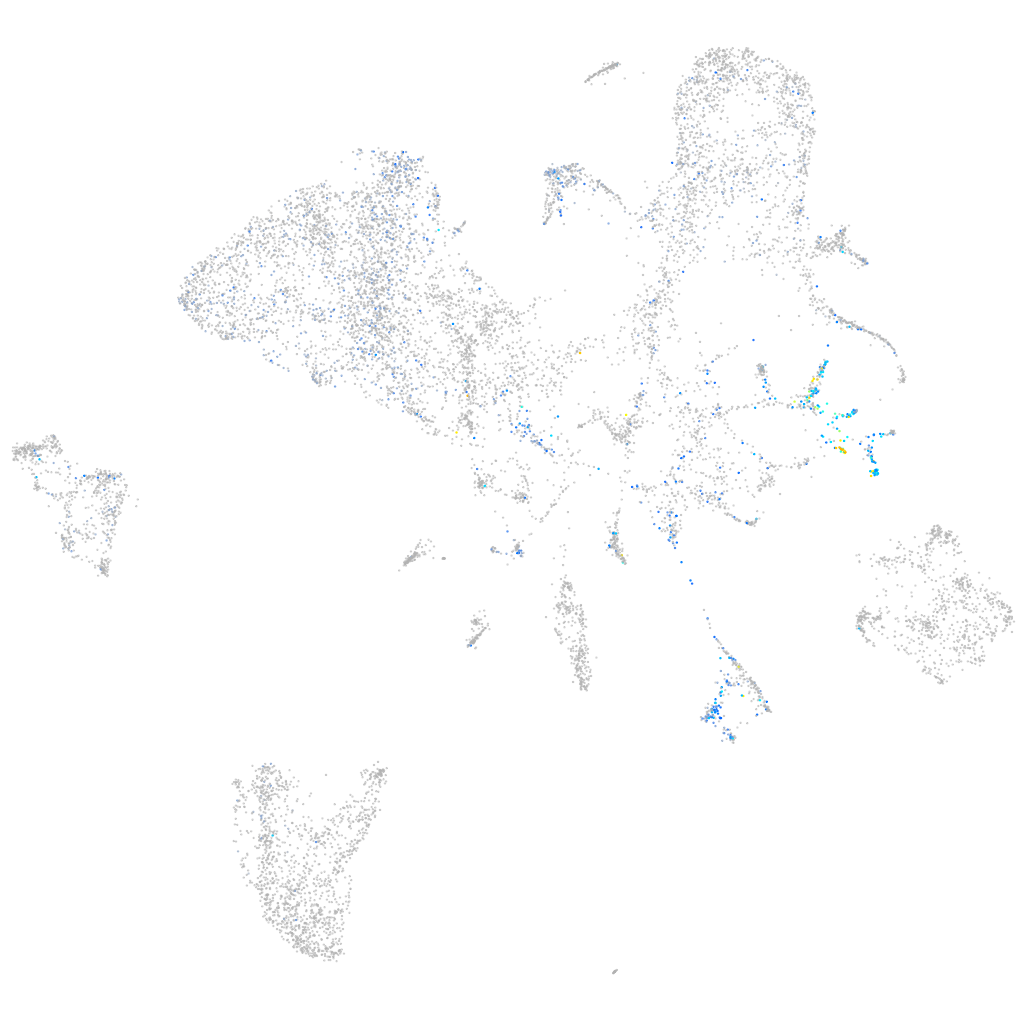
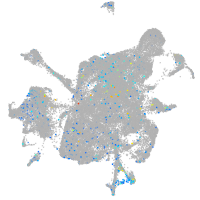
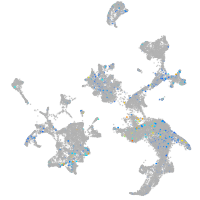
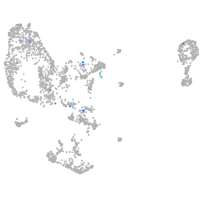
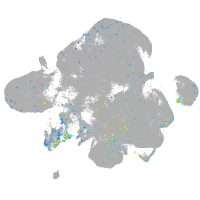
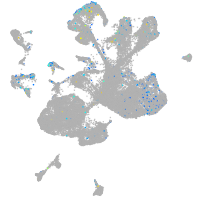
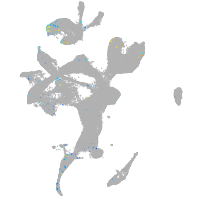
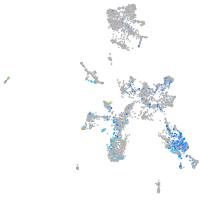
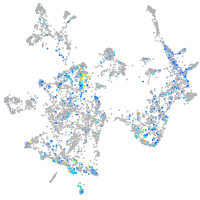
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.489 | gapdh | -0.116 |
| ndufa4l2a | 0.478 | gatm | -0.111 |
| ins | 0.463 | ahcy | -0.109 |
| kcnj11 | 0.437 | gamt | -0.101 |
| pcsk2 | 0.417 | hdlbpa | -0.091 |
| SLC5A10 | 0.407 | bhmt | -0.088 |
| si:dkey-153k10.9 | 0.405 | ela2l | -0.084 |
| rprmb | 0.396 | CELA1 (1 of many) | -0.084 |
| pax6b | 0.396 | zgc:112160 | -0.084 |
| scg3 | 0.389 | apoeb | -0.084 |
| slc30a2 | 0.386 | prss1 | -0.083 |
| isl1 | 0.384 | prss59.1 | -0.083 |
| neurod1 | 0.372 | cpb1 | -0.082 |
| ptn | 0.367 | ctrb1 | -0.081 |
| rasd4 | 0.364 | ela3l | -0.081 |
| LOC100537384 | 0.356 | fep15 | -0.081 |
| pcsk1nl | 0.353 | ela2 | -0.081 |
| gpr142 | 0.347 | cpa5 | -0.081 |
| tspan7b | 0.347 | spink4 | -0.081 |
| pcsk1 | 0.335 | cel.1 | -0.081 |
| scg2a | 0.329 | prss59.2 | -0.081 |
| vat1 | 0.328 | pdia2 | -0.081 |
| rims2a | 0.325 | apoc2 | -0.081 |
| syt1a | 0.323 | apoc1 | -0.080 |
| mir7a-1 | 0.322 | sycn.2 | -0.079 |
| fam49bb | 0.319 | erp27 | -0.079 |
| insm1a | 0.313 | zgc:136461 | -0.079 |
| gpr22a | 0.309 | cel.2 | -0.079 |
| gcgb | 0.309 | fbp1b | -0.079 |
| camk1da | 0.305 | cpa4 | -0.078 |
| foxo1a | 0.301 | si:ch211-240l19.5 | -0.078 |
| kcnk9 | 0.293 | c6ast4 | -0.078 |
| CR774179.5 | 0.291 | mat1a | -0.077 |
| vwa5b2 | 0.290 | aldob | -0.077 |
| pdx1 | 0.288 | slc16a10 | -0.077 |