abhydrolase domain containing 10b
ZFIN 
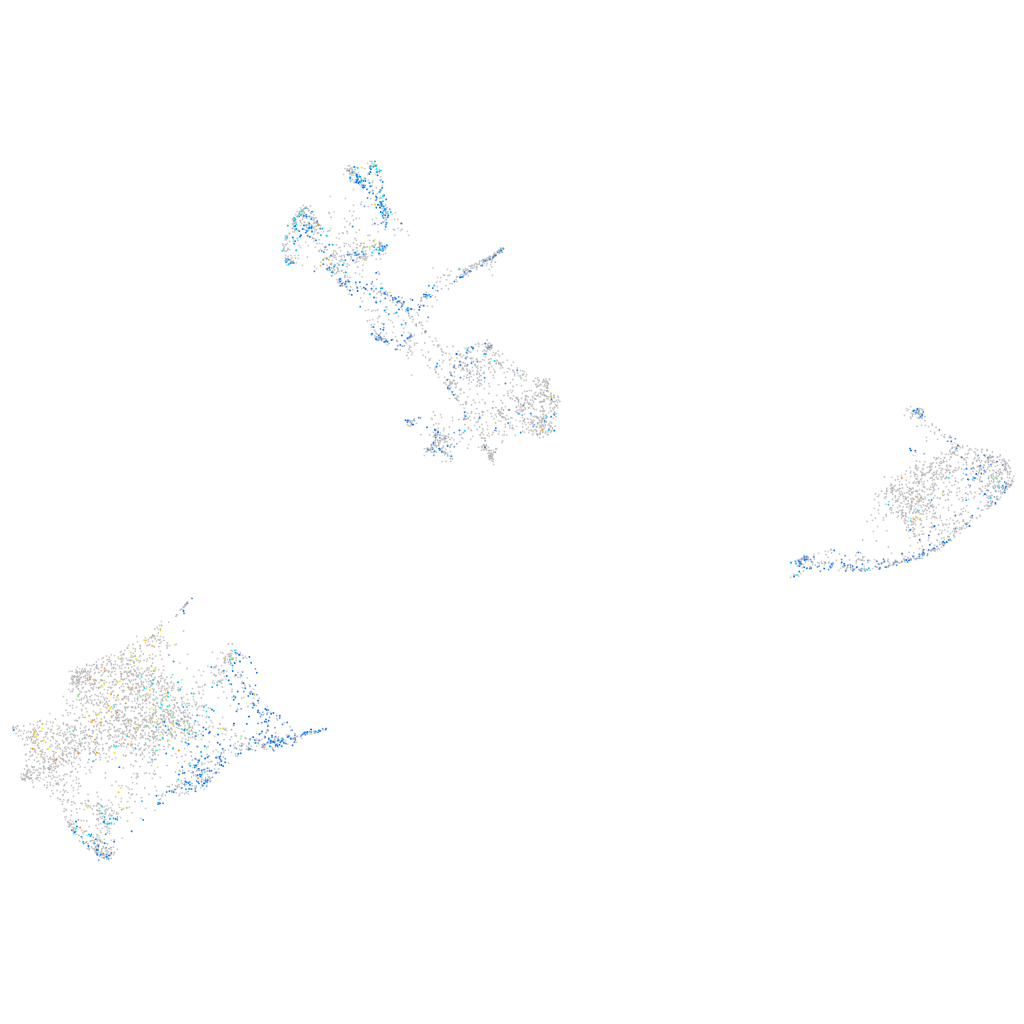
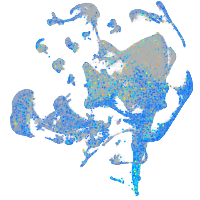
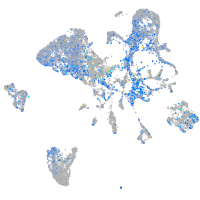
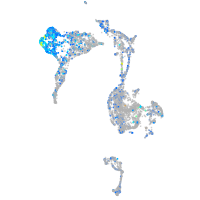
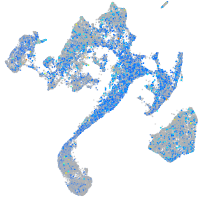
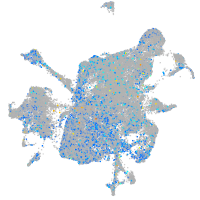
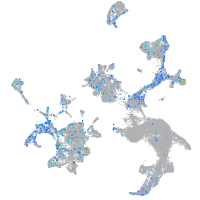
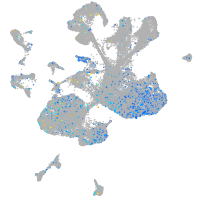
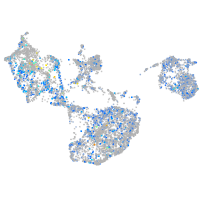
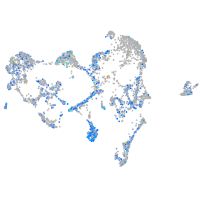
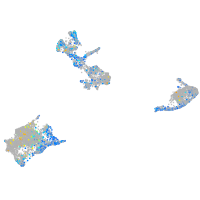
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| impdh1b | 0.164 | icn | -0.115 |
| pnp4a | 0.160 | s100a10b | -0.100 |
| tagln3b | 0.159 | tyrp1b | -0.088 |
| cmtm6 | 0.158 | tuba1c | -0.087 |
| inka1a | 0.158 | pmela | -0.080 |
| tfec | 0.157 | tyrp1a | -0.079 |
| adsl | 0.153 | zgc:91968 | -0.078 |
| phgdh | 0.149 | stmn1b | -0.077 |
| glulb | 0.148 | gpm6aa | -0.077 |
| gmps | 0.147 | prkar1b | -0.076 |
| paics | 0.146 | hbae3 | -0.074 |
| prps1a | 0.143 | oca2 | -0.074 |
| guk1a | 0.141 | epb41a | -0.072 |
| apoda.1 | 0.139 | tmsb | -0.072 |
| agtrap | 0.139 | dct | -0.071 |
| unm-sa821 | 0.138 | pvalb1 | -0.069 |
| psat1 | 0.137 | ndrg4 | -0.068 |
| sytl2b | 0.137 | nova2 | -0.067 |
| tpd52l1 | 0.136 | anxa1a | -0.067 |
| zgc:110239 | 0.136 | mylpfa | -0.064 |
| prelid3b | 0.135 | pvalb2 | -0.064 |
| si:dkey-197i20.6 | 0.134 | elavl3 | -0.064 |
| alx4a | 0.134 | ckbb | -0.063 |
| psph | 0.134 | actc1b | -0.062 |
| si:ch211-256m1.8 | 0.133 | gng3 | -0.062 |
| defbl1 | 0.131 | slc24a5 | -0.061 |
| akap12a | 0.129 | tuba1a | -0.061 |
| si:ch73-89b15.3 | 0.129 | tmsb2 | -0.061 |
| sod1 | 0.129 | sncb | -0.060 |
| smim29 | 0.128 | mtbl | -0.060 |
| pltp | 0.128 | ptmaa | -0.058 |
| sort1a | 0.128 | myhz1.1 | -0.058 |
| nucb2b | 0.127 | snap25b | -0.058 |
| tcirg1a | 0.127 | scrt2 | -0.058 |
| alx4b | 0.127 | zc4h2 | -0.057 |