"ATP-binding cassette, sub-family C (CFTR/MRP), member 5"
ZFIN 
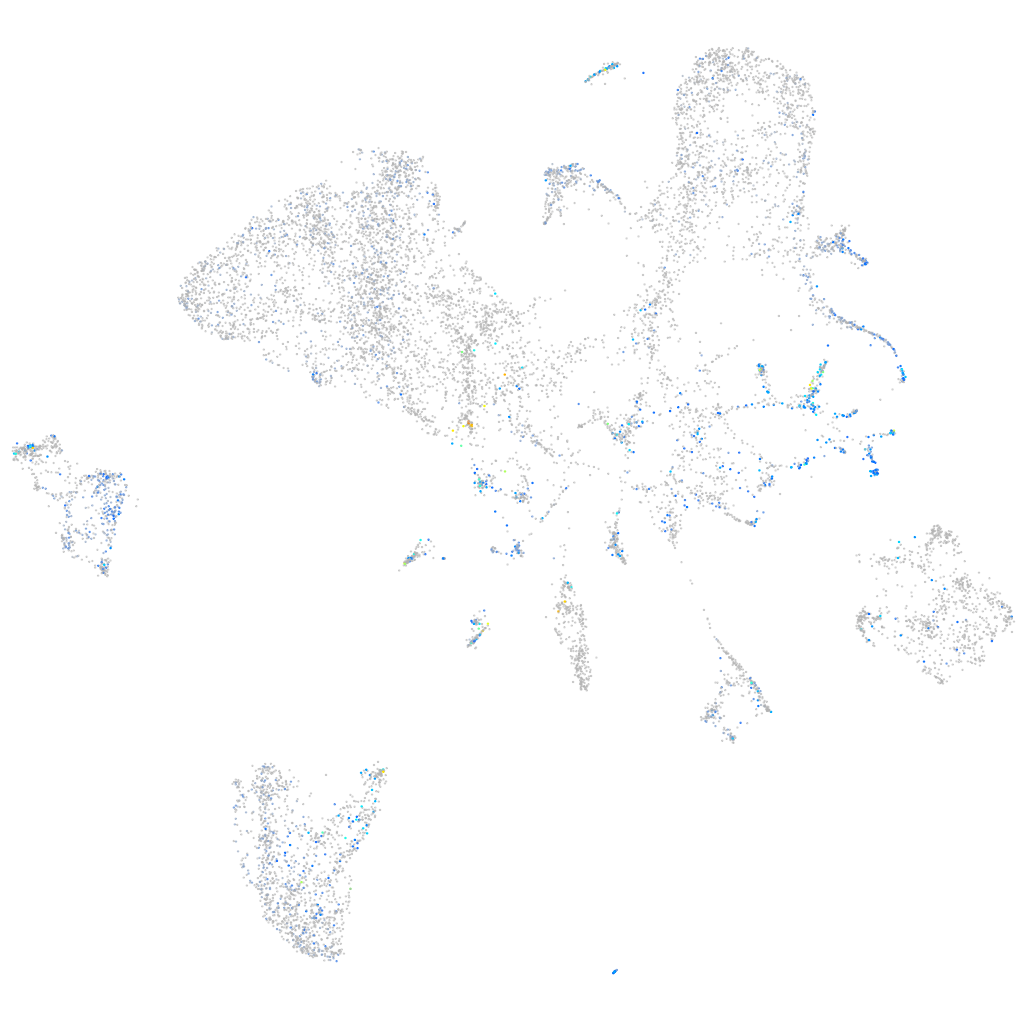
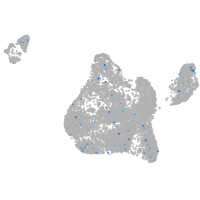
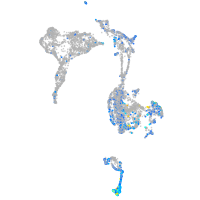
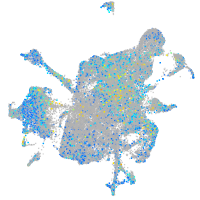
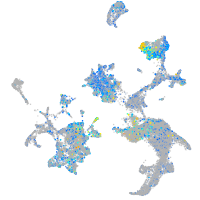
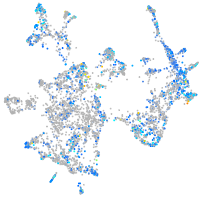
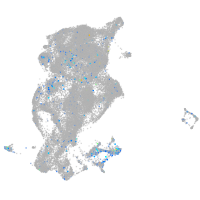
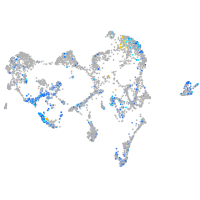
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.236 | gstt1a | -0.160 |
| CR383676.1 | 0.233 | gamt | -0.159 |
| neurod1 | 0.232 | ahcy | -0.158 |
| syt1a | 0.221 | hspe1 | -0.153 |
| scgn | 0.214 | apoc2 | -0.152 |
| pcsk1nl | 0.210 | gcshb | -0.151 |
| cplx2 | 0.203 | suclg1 | -0.150 |
| vamp2 | 0.200 | apoa4b.1 | -0.150 |
| gapdhs | 0.198 | afp4 | -0.149 |
| rnasekb | 0.198 | fbp1b | -0.148 |
| calm1b | 0.198 | gapdh | -0.147 |
| nrsn1 | 0.193 | scp2a | -0.139 |
| pax6b | 0.193 | gstr | -0.139 |
| vat1 | 0.189 | si:dkey-16p21.8 | -0.139 |
| mir7a-1 | 0.188 | aldob | -0.139 |
| ier2b | 0.184 | msrb2 | -0.132 |
| atp6v0cb | 0.182 | shmt1 | -0.131 |
| insm1a | 0.182 | suclg2 | -0.131 |
| c2cd4a | 0.182 | apoa1b | -0.131 |
| sat1a.2 | 0.181 | glud1b | -0.129 |
| pcsk1 | 0.180 | eno3 | -0.129 |
| hepacam2 | 0.177 | gpx4a | -0.129 |
| egr4 | 0.174 | sult2st2 | -0.128 |
| LOC100537384 | 0.173 | rdh1 | -0.127 |
| gpc1a | 0.172 | gatm | -0.126 |
| id4 | 0.171 | nipsnap3a | -0.125 |
| isl1 | 0.169 | cox6b1 | -0.124 |
| zgc:101731 | 0.168 | pgm1 | -0.123 |
| ccni | 0.168 | mat1a | -0.123 |
| dusp2 | 0.167 | cdo1 | -0.123 |
| pcsk2 | 0.165 | COX5B | -0.123 |
| scg2a | 0.162 | ftcd | -0.123 |
| scg2b | 0.161 | apoc1 | -0.122 |
| aldocb | 0.161 | agxta | -0.122 |
| si:dkey-153k10.9 | 0.161 | phyhd1 | -0.122 |