arylacetamide deacetylase-like 4
ZFIN 
Other cell groups


all cells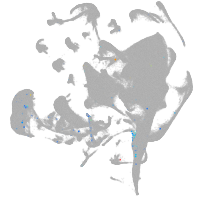 |
blastomeres |
PGCs |
endoderm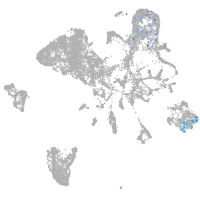 |
axial mesoderm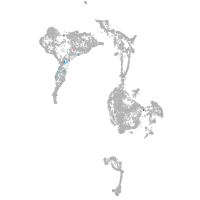 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 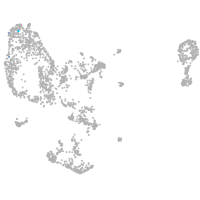 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cpo | 0.117 | marcksl1a | -0.026 |
| si:ch211-77g15.32 | 0.117 | tuba1c | -0.025 |
| si:dkey-30c15.13 | 0.116 | fabp3 | -0.023 |
| zanl | 0.116 | gpm6aa | -0.023 |
| ace2 | 0.115 | nova2 | -0.022 |
| aqp8a.2 | 0.114 | rtn1a | -0.022 |
| chs1 | 0.114 | stmn1b | -0.021 |
| LOC103908636 | 0.113 | atp6v0cb | -0.020 |
| mep1a.2 | 0.113 | elavl3 | -0.020 |
| wu:fa56d06 | 0.113 | fam168a | -0.020 |
| anpepa | 0.111 | mdkb | -0.020 |
| slc6a19a.2 | 0.111 | celf2 | -0.019 |
| vwa11 | 0.111 | jpt1b | -0.019 |
| zgc:153968 | 0.111 | rnasekb | -0.019 |
| AL831745.1 | 0.110 | gapdhs | -0.018 |
| adgrg4a | 0.109 | gpm6ab | -0.018 |
| mbl2 | 0.108 | hbbe1.3 | -0.018 |
| cyp2x9 | 0.107 | hmgb3a | -0.018 |
| enpp7.1 | 0.106 | sncb | -0.018 |
| ptprh | 0.106 | tuba1a | -0.018 |
| IZUMO1R | 0.105 | vdac1 | -0.018 |
| LOC100536156 | 0.105 | cspg5a | -0.017 |
| pcxb | 0.105 | gng3 | -0.017 |
| pdzd3b | 0.105 | hbae3 | -0.017 |
| aoc1 | 0.105 | hmgb1b | -0.017 |
| btr05 | 0.104 | vamp2 | -0.017 |
| ca4b | 0.104 | atpv0e2 | -0.016 |
| si:ch211-133l5.7 | 0.104 | elavl4 | -0.016 |
| slc6a19b | 0.104 | hbae1.1 | -0.016 |
| zgc:77748 | 0.104 | rtn1b | -0.016 |
| sdsl | 0.103 | tmsb | -0.016 |
| slc5a1 | 0.103 | tubb5 | -0.016 |
| clrn3 | 0.101 | cadm3 | -0.015 |
| si:dkey-283b1.6 | 0.101 | ckbb | -0.015 |
| zgc:158846 | 0.101 | dbn1 | -0.015 |




