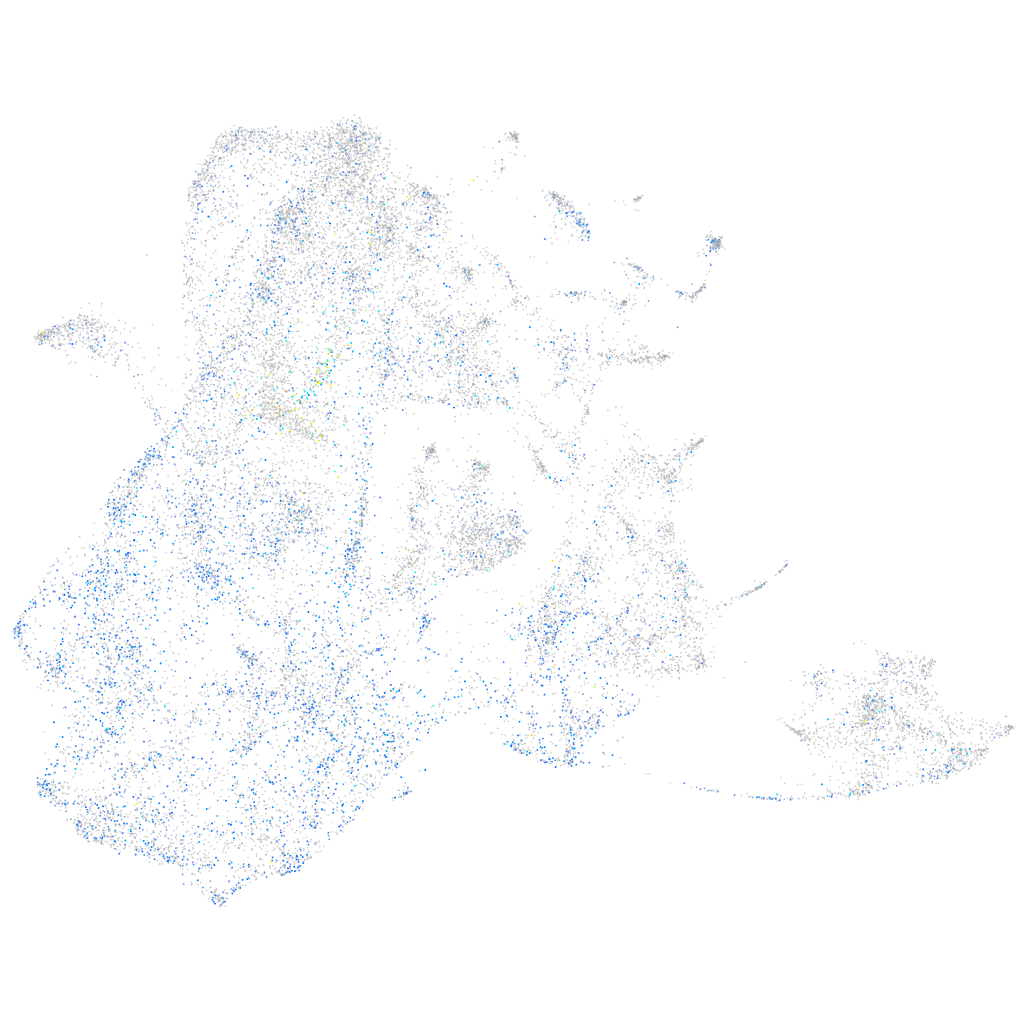
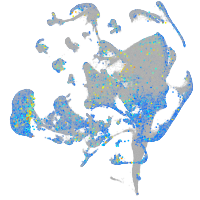
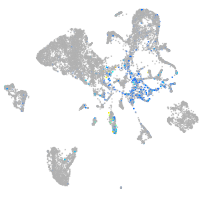
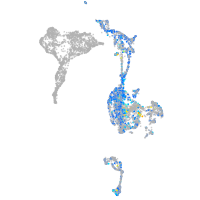
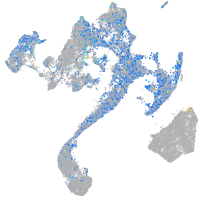
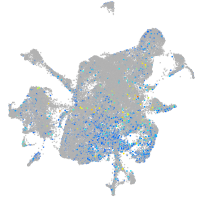
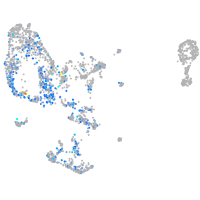
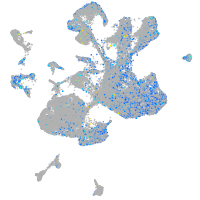
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| NC-002333.4 | 0.249 | ahcy | -0.152 |
| CABZ01072614.1 | 0.246 | nme2b.1 | -0.151 |
| NC-002333.17 | 0.217 | ucp2 | -0.146 |
| fkbp9 | 0.201 | si:dkey-33c14.3 | -0.146 |
| asph | 0.182 | eef1da | -0.140 |
| LOC110439627 | 0.167 | cxl34b.11 | -0.140 |
| col14a1a | 0.166 | krt91 | -0.140 |
| krt8 | 0.162 | epgn | -0.138 |
| p4ha1b | 0.157 | eef1b2 | -0.130 |
| p3h1 | 0.156 | sod1 | -0.127 |
| LOC110439372 | 0.155 | gapdhs | -0.127 |
| fbn2b | 0.152 | zgc:136930 | -0.124 |
| mdka | 0.149 | si:dkey-183i3.5 | -0.121 |
| plod3 | 0.145 | mcl1a | -0.121 |
| tpm4a | 0.144 | apoa2 | -0.121 |
| rrbp1b | 0.143 | rhbg | -0.117 |
| crtap | 0.141 | pvalb1 | -0.116 |
| fermt1 | 0.141 | romo1 | -0.112 |
| dag1 | 0.141 | tdh | -0.108 |
| igf2bp1 | 0.138 | zgc:77439 | -0.107 |
| tram1 | 0.135 | s100v2 | -0.106 |
| nid2a | 0.135 | rplp2 | -0.101 |
| kdelr2b | 0.133 | prdx2 | -0.101 |
| tinagl1 | 0.132 | krtt1c19e | -0.101 |
| nr2f5 | 0.132 | mylipb | -0.100 |
| ssr3 | 0.132 | eif4a1b | -0.099 |
| akap12b | 0.130 | prelid3b | -0.099 |
| myl6 | 0.130 | pvalb2 | -0.099 |
| ppib | 0.128 | igfbp7 | -0.097 |
| emp2 | 0.128 | eif4ebp3 | -0.097 |
| fkbp10b | 0.128 | lgals1l1 | -0.097 |
| sec61b | 0.127 | apoa1b | -0.096 |
| bcam | 0.125 | znf706 | -0.096 |
| CABZ01075068.1 | 0.124 | crip1 | -0.096 |
| myh9a | 0.123 | atp5mc3b | -0.095 |