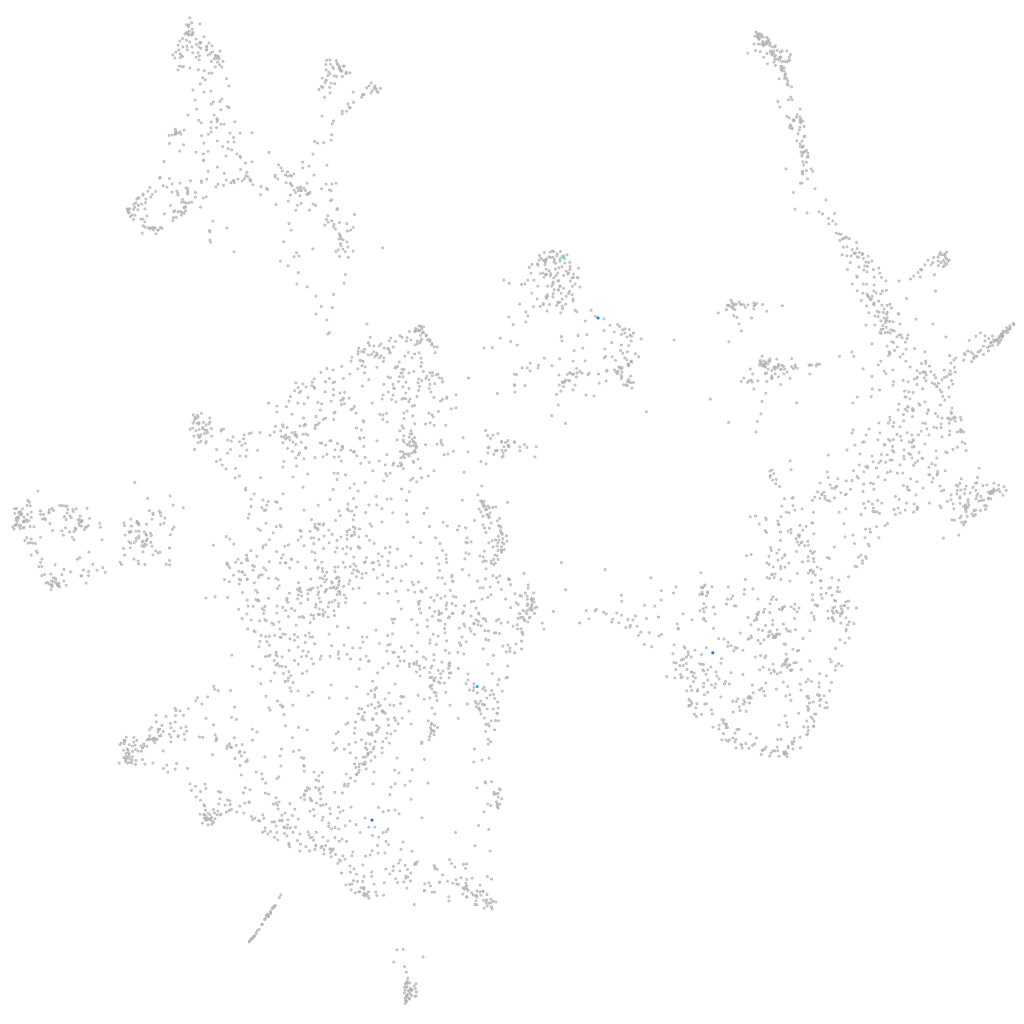
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 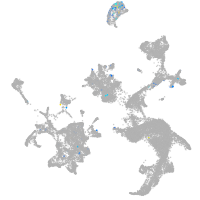 |
pronephros  |
neural |
spinal cord / glia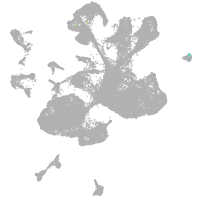 |
eye |
taste / olfactory |
otic / lateral line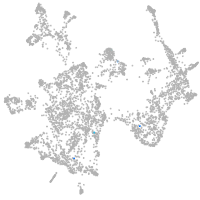 |
epidermis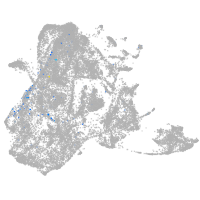 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR388166.1 | 0.660 | rbx1 | -0.040 |
| zte38 | 0.616 | rps26l | -0.039 |
| slc6a4b | 0.590 | rpl10 | -0.037 |
| si:dkey-33c9.6 | 0.557 | tuba8l4 | -0.036 |
| LOC110439438 | 0.544 | cox7c | -0.035 |
| fndc5a | 0.451 | rps5 | -0.034 |
| si:ch211-59d15.4 | 0.412 | atp5pb | -0.033 |
| ppef1 | 0.409 | snrpg | -0.033 |
| rap1gap2b | 0.402 | eif3f | -0.032 |
| XLOC-019059 | 0.401 | csnk2b | -0.031 |
| CU928046.1 | 0.387 | cox8a | -0.031 |
| hrh3 | 0.386 | rpl23a | -0.030 |
| gas2l2 | 0.378 | sf3b5 | -0.030 |
| CT027980.1 | 0.363 | snw1 | -0.030 |
| kcng3 | 0.357 | mt-nd1 | -0.029 |
| slco1f1 | 0.350 | gtf2f2a | -0.029 |
| pimr128 | 0.350 | cct2 | -0.029 |
| nkx2.4a | 0.342 | eif2s2 | -0.029 |
| CRACR2A | 0.341 | atp5pd | -0.028 |
| tmem121ab | 0.341 | eif3ha | -0.028 |
| XLOC-026855 | 0.336 | ndufb8 | -0.028 |
| calcr | 0.332 | cbx3a | -0.028 |
| syngap1a | 0.328 | ddx61 | -0.028 |
| tac1 | 0.318 | trappc1 | -0.028 |
| cnn1b | 0.311 | ndufab1a | -0.028 |
| frs3 | 0.306 | selenot1a | -0.028 |
| tnni1d | 0.306 | COX5B | -0.027 |
| glis1a | 0.304 | pbdc1 | -0.027 |
| dkk3a | 0.300 | ndufa5 | -0.027 |
| nkx2.4b | 0.295 | ndufs5 | -0.027 |
| CABZ01086574.1 | 0.292 | psmd14 | -0.027 |
| ccm2l | 0.286 | pfdn6 | -0.027 |
| prdm13 | 0.285 | ptges3b | -0.026 |
| TMEM114 | 0.280 | mtdha | -0.026 |
| nkx2.2a | 0.279 | psmd8 | -0.026 |