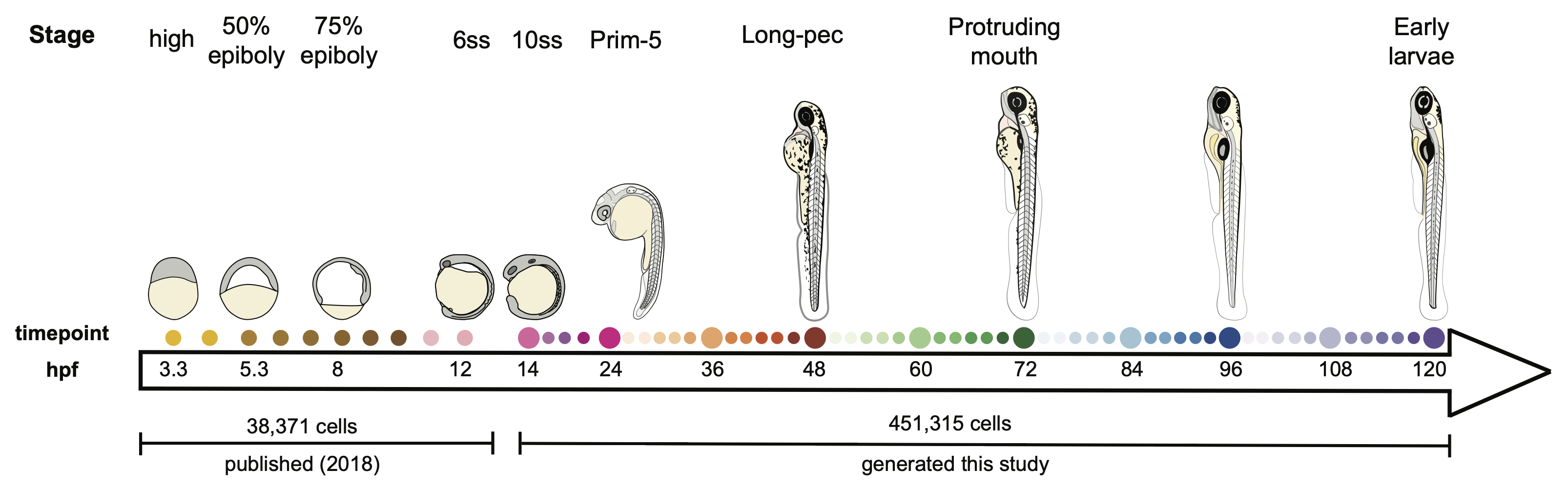
Daniocell is an online resource that presents single-cell gene expression data generated from whole-animal wild-type (TL/AB) zebrafish embryos and larvae across 62 stages of development, spanning 3.3–120 hours post-fertilization. This combines data published in "Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis" (2018) and "Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development" (2023). This site is work from the Farrell Lab at NICHD. If you use it in your work, please cite it!
Using Daniocell
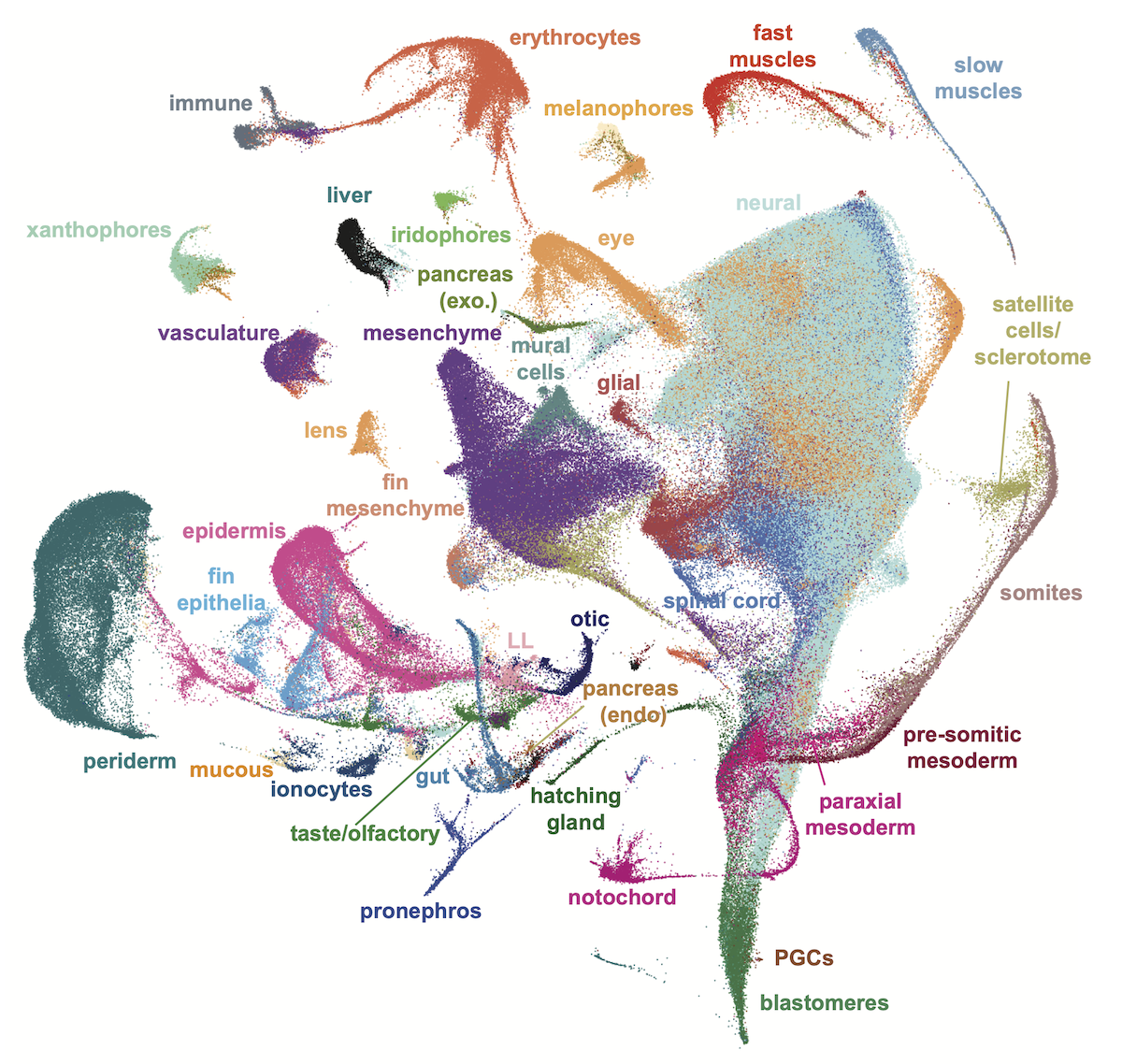
Pages in Daniocell are oriented around either a gene, or a cluster identified within our study. The data was first analyzed as a whole-animal data set, and these results are presented on the "all cells" pages. Cells were then assigned to 19 broad tissue types, based on cluster identity, transcriptional similarity, developmental stage, and established literature. Within each of these, the data were reanalyzed, and the results of those tissue-specific analyses can be viewed by clicking a specific tissue on the right of a gene page.
On gene pages, expression can be viewed either within the animal or tissue-specific UMAP projections (mouse over the UMAP to help orient yourself about which cluster you're looking at) or within time-resolved dot plots. Additionally, correlation tables identify genes with the most similar or dissimilar expression patterns, either across all cells, or within individual tissues.
Cluster pages can be reached either through the search bar, or by clicking on a specific cell type from the dot plot of a tissue-specific gene expression page. Cluster pages identify when cells of a given cluster are present, proliferating, and in 'long-term' transcriptional states, and also identify marker genes of each cluster (both 'most specific' and 'most highly expressed'.)
For more information about the analyses presented in each section on the website, mouse over the (?) symbol, which will then display additional information.
Data Availability
In addition to browsing the data online through Daniocell, it can be obtained in additional formats for reanalysis:
- The raw sequencing reads in FASTQ format can be obtained from NCBI GEO GSE223922.
- A UMI counts table containing all cells can also be obtained from NCBI GEO GSE223922.
- The processed Seurat (v4) object:
- Daniocell2023_SeuratV4.rds (Seurat object)
- cluster_annotations.csv (Cluster annotations)
- daniocell_load.R (R script to load object and annotations, with function to regenerate tissue-specific cropped objects)
Annotation Amendments
As those of you who have worked with single-cell RNAseq data will be familiar, annotating cell identities can be a painstaking and difficult process. We expect that researchers who are more expert in particular tissues or cell types may wish to suggest improvements or amendments to the annotations that we have produced, and we gladly welcome the feedback. Our goal is to incorporate these suggestions on a regular schedule and update the information on this site, including acknowledgment of those researchers who have contributed.
To facilitate this, please email Jeffrey Farrell and Abhinav Sur with the following information:
- Current annotation and cluster ID of cells
- Is the cluster completely misidentified? Or is the cluster a mixture of multiple cell types, and a subset of them would ideally be assigned a different identity?
- The expression characteristics that your amendment is based on:
- The expressed genes that your identification is based on
- Any expressed genes that disagree with the current annotation
- When possible, references from the literature and/or ZFIN to support the amendment you propose
- When possible, the closest equivalent from the ZFIN Anatomy / Cell type ontology
- Would you like to be attributed for your contribution? If so, please
indicate:
- The name(s) you would like the amendment attributed to
- (Optional) Your ORCid
- Any other information you think would be helpful to us or would add additional clarification
Acknowledgments
This website would not have been possible without the generous investment and support of the NICHD Research Informatics Support Division, including Loc Vu (for help with Javascript programming, including the search bar), Matt Breymaier (for help with testing and managing online access), Nicki Swan (for help with graphic design), and Ryan Dale (for help with overall design and implementation).
Version History
- 2023/09/20 (v1.1): Amended persistence measurements, annotations (3 altered labels), improved search, and Seurat object availability
- 2023/03/21 (v1.0): Public release
- 2023/03/07 (v0.2): Beta release 2 (NICHD colleagues)
- 2022/09/25 (v0.1): Beta release 1 (NICHD colleagues)